| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
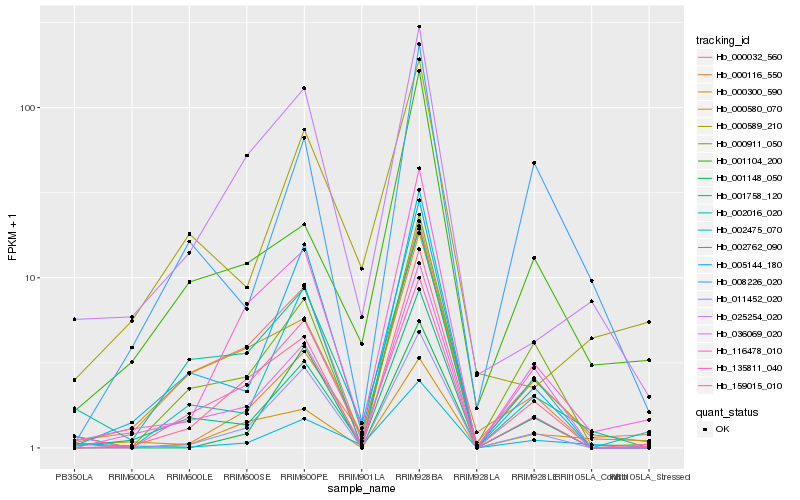
| 1 |
Hb_002762_090 |
0.0 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_116478_010 |
0.1766091481 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF003-like [Jatropha curcas] |
| 3 |
Hb_000300_590 |
0.1890889501 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-11-like [Jatropha curcas] |
| 4 |
Hb_001758_120 |
0.198247486 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_025254_020 |
0.2048912989 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Vitis vinifera] |
| 6 |
Hb_002475_070 |
0.2078274471 |
- |
- |
hypothetical protein POPTR_0017s02440g, partial [Populus trichocarpa] |
| 7 |
Hb_002016_020 |
0.2086508935 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 8 |
Hb_000032_560 |
0.2093619499 |
- |
- |
hypothetical protein JCGZ_15037 [Jatropha curcas] |
| 9 |
Hb_001104_200 |
0.2110950233 |
- |
- |
Phytosulfokine 3 precursor, putative [Theobroma cacao] |
| 10 |
Hb_001148_050 |
0.216183031 |
- |
- |
- |
| 11 |
Hb_159015_010 |
0.2172985092 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 12 |
Hb_000589_210 |
0.2173617125 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |
| 13 |
Hb_135811_040 |
0.2177436922 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_036069_020 |
0.2182751663 |
- |
- |
PREDICTED: receptor protein kinase-like protein At4g34220 [Jatropha curcas] |
| 15 |
Hb_000580_070 |
0.2189716587 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 16 |
Hb_008226_020 |
0.2191766325 |
- |
- |
hypothetical protein JCGZ_03321 [Jatropha curcas] |
| 17 |
Hb_005144_180 |
0.2197370164 |
- |
- |
PREDICTED: protein enabled homolog [Populus euphratica] |
| 18 |
Hb_000911_050 |
0.2248777087 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 21 [Jatropha curcas] |
| 19 |
Hb_011452_020 |
0.2255077594 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 4 [Jatropha curcas] |
| 20 |
Hb_000116_550 |
0.2273331799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |