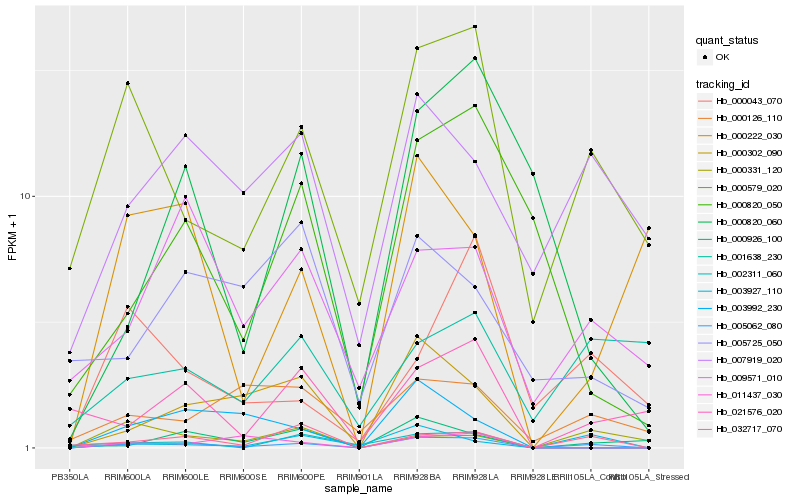
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005062_080 |
0.0 |
- |
- |
MLO-like protein 4 [Gossypium arboreum] |
| 2 |
Hb_000579_020 |
0.2560101641 |
- |
- |
hypothetical protein POPTR_0017s06850g [Populus trichocarpa] |
| 3 |
Hb_032717_070 |
0.257125902 |
- |
- |
PREDICTED: mucin-2-like [Jatropha curcas] |
| 4 |
Hb_003927_110 |
0.292789571 |
- |
- |
hypothetical protein PRUPE_ppa001478mg [Prunus persica] |
| 5 |
Hb_007919_020 |
0.3070480875 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 6 |
Hb_000926_100 |
0.3117575423 |
- |
- |
hypothetical protein B456_007G076800 [Gossypium raimondii] |
| 7 |
Hb_000302_090 |
0.3149561636 |
- |
- |
PREDICTED: protein SGT1 homolog [Prunus mume] |
| 8 |
Hb_009571_010 |
0.3176196498 |
- |
- |
early-responsive to dehydration family protein [Populus trichocarpa] |
| 9 |
Hb_021576_020 |
0.3218712186 |
- |
- |
PREDICTED: uncharacterized protein LOC104426132 [Eucalyptus grandis] |
| 10 |
Hb_003992_230 |
0.3236252227 |
- |
- |
PREDICTED: uncharacterized protein LOC105634858 [Jatropha curcas] |
| 11 |
Hb_000820_050 |
0.3262400998 |
- |
- |
hypothetical protein EUGRSUZ_H02626 [Eucalyptus grandis] |
| 12 |
Hb_000126_110 |
0.3305648369 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X3 [Populus euphratica] |
| 13 |
Hb_000820_060 |
0.3390953935 |
- |
- |
- |
| 14 |
Hb_011437_030 |
0.339671382 |
- |
- |
PREDICTED: uncharacterized protein LOC105037131, partial [Elaeis guineensis] |
| 15 |
Hb_000043_070 |
0.3427039337 |
- |
- |
PREDICTED: transcription factor IBH1 [Jatropha curcas] |
| 16 |
Hb_000222_030 |
0.3444016071 |
- |
- |
- |
| 17 |
Hb_002311_060 |
0.3474348322 |
- |
- |
PREDICTED: protein trichome birefringence-like 4 [Jatropha curcas] |
| 18 |
Hb_000331_120 |
0.3522343675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001638_230 |
0.3564922822 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 20 |
Hb_005725_050 |
0.3591763508 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |