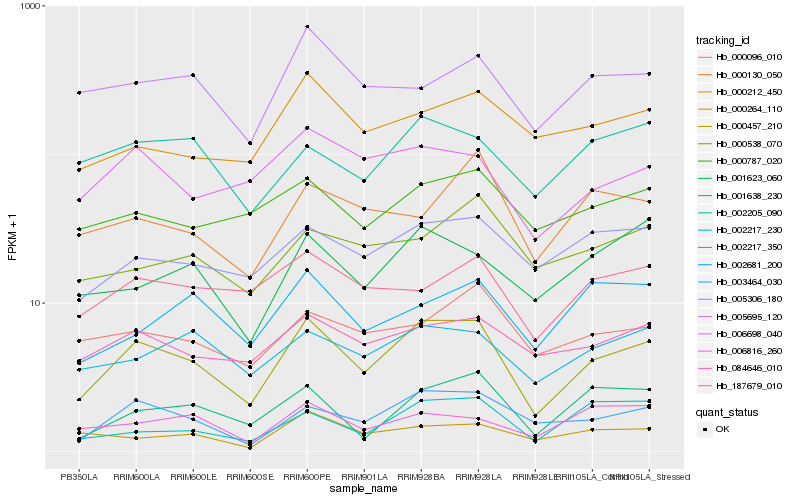
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000457_210 |
0.0 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 2 |
Hb_003464_030 |
0.1310911503 |
- |
- |
PREDICTED: 60S ribosomal protein L24-like [Jatropha curcas] |
| 3 |
Hb_005306_180 |
0.1527651881 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_006698_040 |
0.1559988559 |
- |
- |
bHLH1 transcription factor [Hevea brasiliensis] |
| 5 |
Hb_084646_010 |
0.1560478011 |
- |
- |
hypothetical protein CICLE_v10031746mg [Citrus clementina] |
| 6 |
Hb_002217_350 |
0.1562010952 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 7 |
Hb_001638_230 |
0.1577369531 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 8 |
Hb_001623_060 |
0.1632628885 |
- |
- |
potassium channel beta, putative [Ricinus communis] |
| 9 |
Hb_002205_090 |
0.1646088457 |
- |
- |
RecName: Full=Superoxide dismutase [Mn], mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 10 |
Hb_005695_120 |
0.1657582486 |
- |
- |
Pollen-specific protein C13 precursor, putative [Ricinus communis] |
| 11 |
Hb_187679_010 |
0.1658290179 |
- |
- |
- |
| 12 |
Hb_000787_020 |
0.1662284725 |
- |
- |
PREDICTED: glucose-6-phosphate/phosphate translocator 1, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_000096_010 |
0.1664823317 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Citrus sinensis] |
| 14 |
Hb_000212_450 |
0.1687609112 |
- |
- |
PREDICTED: basic leucine zipper and W2 domain-containing protein 2 [Jatropha curcas] |
| 15 |
Hb_006816_260 |
0.170183502 |
- |
- |
hypothetical protein JCGZ_23244 [Jatropha curcas] |
| 16 |
Hb_002681_200 |
0.1702041572 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2 [Jatropha curcas] |
| 17 |
Hb_000264_110 |
0.1716914489 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 13 [Jatropha curcas] |
| 18 |
Hb_002217_230 |
0.172930541 |
- |
- |
PREDICTED: uncharacterized protein LOC105643583 [Jatropha curcas] |
| 19 |
Hb_000538_070 |
0.1749809491 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 20 |
Hb_000130_050 |
0.1761633726 |
- |
- |
14-3-3 protein, putative [Ricinus communis] |