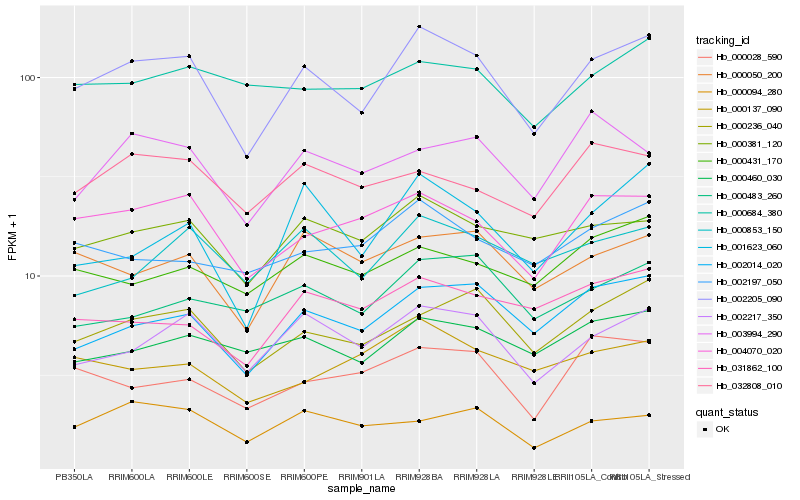
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002205_090 |
0.0 |
- |
- |
RecName: Full=Superoxide dismutase [Mn], mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 2 |
Hb_002217_350 |
0.0874355174 |
- |
- |
PREDICTED: importin subunit alpha-4-like [Jatropha curcas] |
| 3 |
Hb_000236_040 |
0.0909770475 |
- |
- |
PREDICTED: uncharacterized protein LOC105633150 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004070_020 |
0.0910548619 |
- |
- |
rhythmically-expressed protein 2 protein, putative [Ricinus communis] |
| 5 |
Hb_002014_020 |
0.0913648332 |
- |
- |
PREDICTED: protein Mpv17 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000381_120 |
0.0976688883 |
- |
- |
PREDICTED: uncharacterized protein LOC105648175 [Jatropha curcas] |
| 7 |
Hb_000050_200 |
0.1035714203 |
- |
- |
ergosterol biosynthetic protein 28 [Jatropha curcas] |
| 8 |
Hb_000460_030 |
0.1082071217 |
- |
- |
4-hydroxybenzoate octaprenyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000853_150 |
0.1119647362 |
- |
- |
Fumarase 1 isoform 2 [Theobroma cacao] |
| 10 |
Hb_001623_060 |
0.1125921073 |
- |
- |
potassium channel beta, putative [Ricinus communis] |
| 11 |
Hb_002197_050 |
0.115305034 |
- |
- |
hypothetical protein CICLE_v10002157mg [Citrus clementina] |
| 12 |
Hb_000094_280 |
0.1168099614 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 13 |
Hb_000431_170 |
0.1168447737 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 511 [Jatropha curcas] |
| 14 |
Hb_031862_100 |
0.1168621321 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_032808_010 |
0.1168639558 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 16 |
Hb_000483_260 |
0.1180658537 |
- |
- |
PREDICTED: dihydroorotase, mitochondrial isoform X1 [Populus euphratica] |
| 17 |
Hb_000684_380 |
0.1183753826 |
- |
- |
PREDICTED: 60S ribosomal protein L4 [Vitis vinifera] |
| 18 |
Hb_000028_590 |
0.1185957012 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Jatropha curcas] |
| 19 |
Hb_003994_290 |
0.1186764704 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase GAPCP2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000137_090 |
0.1187852029 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Vitis vinifera] |