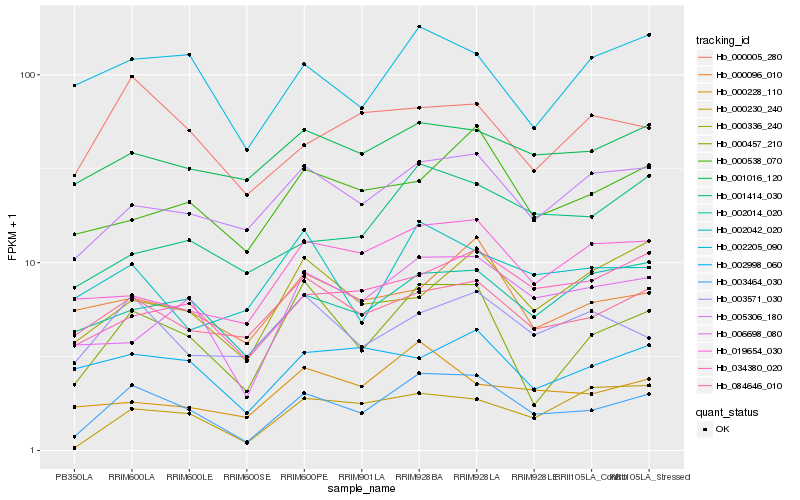
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003464_030 |
0.0 |
- |
- |
PREDICTED: 60S ribosomal protein L24-like [Jatropha curcas] |
| 2 |
Hb_000457_210 |
0.1310911503 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 3 |
Hb_005306_180 |
0.1612968065 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_006698_080 |
0.1625629302 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_084646_010 |
0.1675688314 |
- |
- |
hypothetical protein CICLE_v10031746mg [Citrus clementina] |
| 6 |
Hb_003571_030 |
0.1698959829 |
- |
- |
hypothetical protein JCGZ_25894 [Jatropha curcas] |
| 7 |
Hb_002014_020 |
0.1715239727 |
- |
- |
PREDICTED: protein Mpv17 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000005_280 |
0.1723780767 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 24 homolog 1-like [Citrus sinensis] |
| 9 |
Hb_002205_090 |
0.1727132659 |
- |
- |
RecName: Full=Superoxide dismutase [Mn], mitochondrial; Flags: Precursor [Hevea brasiliensis] |
| 10 |
Hb_034380_020 |
0.174010924 |
- |
- |
PREDICTED: phosphoglycerate mutase-like [Jatropha curcas] |
| 11 |
Hb_000230_240 |
0.1755867819 |
- |
- |
tRNA/rRNA methyltransferase family protein [Populus trichocarpa] |
| 12 |
Hb_000538_070 |
0.1779343406 |
- |
- |
PREDICTED: coatomer subunit delta [Jatropha curcas] |
| 13 |
Hb_002998_060 |
0.1790871257 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 33 homolog [Jatropha curcas] |
| 14 |
Hb_000096_010 |
0.1799845447 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Citrus sinensis] |
| 15 |
Hb_001414_030 |
0.1805250148 |
- |
- |
PREDICTED: aspartate-semialdehyde dehydrogenase [Jatropha curcas] |
| 16 |
Hb_019654_030 |
0.1810404199 |
- |
- |
maoC-like dehydratase domain-containing family protein [Populus trichocarpa] |
| 17 |
Hb_001016_120 |
0.1829689602 |
- |
- |
PREDICTED: 3-dehydroquinate synthase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002042_020 |
0.1831511588 |
- |
- |
hypothetical protein CICLE_v10001869mg [Citrus clementina] |
| 19 |
Hb_000336_240 |
0.1844631188 |
transcription factor |
TF Family: mTERF |
LOC100285792 [Zea mays] |
| 20 |
Hb_000228_110 |
0.1852069884 |
- |
- |
unnamed protein product [Vitis vinifera] |