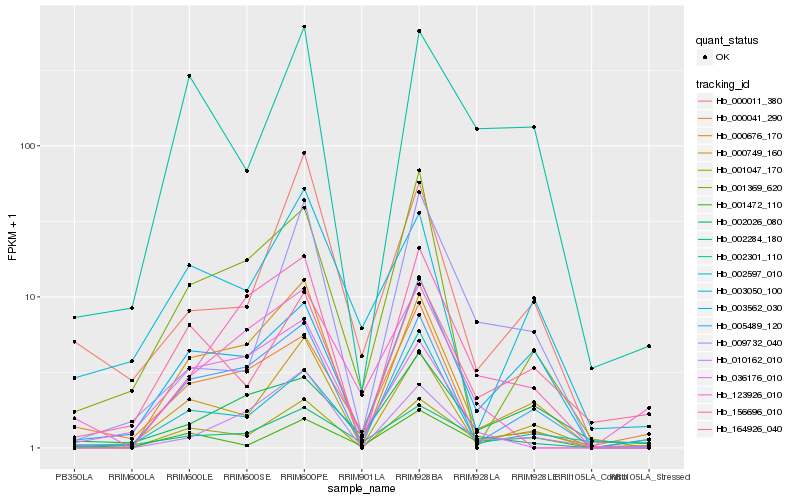
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002284_180 |
0.0 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 2 |
Hb_156696_010 |
0.2097595497 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 3 |
Hb_003562_030 |
0.2162359125 |
- |
- |
- |
| 4 |
Hb_001047_170 |
0.2172598665 |
- |
- |
PREDICTED: uncharacterized protein LOC105649981 [Jatropha curcas] |
| 5 |
Hb_002301_110 |
0.2236985027 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 6 |
Hb_000676_170 |
0.228677276 |
- |
- |
PREDICTED: bifunctional epoxide hydrolase 2 [Jatropha curcas] |
| 7 |
Hb_005489_120 |
0.2290033121 |
- |
- |
leucine rich repeat receptor kinase, putative [Ricinus communis] |
| 8 |
Hb_001472_110 |
0.2293140426 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 9 |
Hb_002026_080 |
0.2312554918 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 10 |
Hb_000041_290 |
0.232992452 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 11 |
Hb_010162_010 |
0.2357882559 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 12 |
Hb_001369_620 |
0.237200531 |
- |
- |
PREDICTED: protein EXORDIUM-like 3 [Jatropha curcas] |
| 13 |
Hb_009732_040 |
0.2379782699 |
- |
- |
hypothetical protein POPTR_0006s04880g [Populus trichocarpa] |
| 14 |
Hb_000749_160 |
0.239074923 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 15 |
Hb_003050_100 |
0.2393734635 |
- |
- |
PREDICTED: uncharacterized protein LOC105633987 [Jatropha curcas] |
| 16 |
Hb_002597_010 |
0.24027836 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Malus domestica] |
| 17 |
Hb_123926_010 |
0.2408443577 |
- |
- |
PREDICTED: probable glutathione peroxidase 8-like [Glycine max] |
| 18 |
Hb_036176_010 |
0.2413343051 |
- |
- |
hypothetical protein CISIN_1g002055mg [Citrus sinensis] |
| 19 |
Hb_164926_040 |
0.243223819 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |
| 20 |
Hb_000011_380 |
0.2434700192 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |