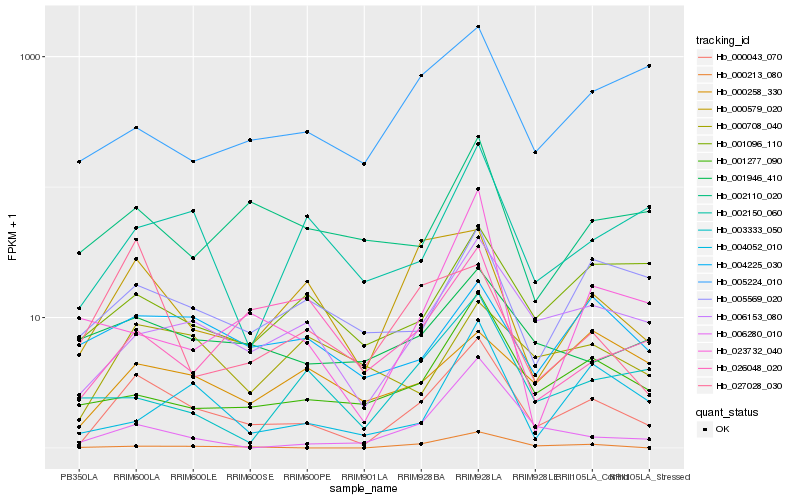
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000043_070 |
0.0 |
- |
- |
PREDICTED: transcription factor IBH1 [Jatropha curcas] |
| 2 |
Hb_006153_080 |
0.2183884215 |
- |
- |
flavin-containing monooxygenase-related family protein [Populus trichocarpa] |
| 3 |
Hb_000579_020 |
0.2452353213 |
- |
- |
hypothetical protein POPTR_0017s06850g [Populus trichocarpa] |
| 4 |
Hb_001277_090 |
0.252307723 |
- |
- |
PREDICTED: uncharacterized protein LOC105632061 [Jatropha curcas] |
| 5 |
Hb_002150_060 |
0.2724646554 |
- |
- |
PREDICTED: arabinogalactan peptide 20-like [Jatropha curcas] |
| 6 |
Hb_005569_020 |
0.2735827623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001946_410 |
0.2790538939 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002110_020 |
0.2844078817 |
- |
- |
PREDICTED: transcription factor bHLH145 [Jatropha curcas] |
| 9 |
Hb_006280_010 |
0.2860300867 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor TOE3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_004225_030 |
0.286706002 |
- |
- |
hypothetical protein F383_15788 [Gossypium arboreum] |
| 11 |
Hb_000213_080 |
0.2886544635 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1-like [Vitis vinifera] |
| 12 |
Hb_000708_040 |
0.2892350213 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004052_010 |
0.2916909649 |
- |
- |
PREDICTED: ninja-family protein AFP3-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_001096_110 |
0.2928284065 |
- |
- |
PREDICTED: probable VAMP-like protein At1g33475 [Jatropha curcas] |
| 15 |
Hb_027028_030 |
0.2956754606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_026048_020 |
0.2980531825 |
- |
- |
PREDICTED: uncharacterized protein LOC105645738 [Jatropha curcas] |
| 17 |
Hb_005224_010 |
0.3014788192 |
- |
- |
glycine-rich protein [Bruguiera gymnorhiza] |
| 18 |
Hb_023732_040 |
0.3015233874 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_000258_330 |
0.301928797 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 20 |
Hb_003333_050 |
0.3021274815 |
- |
- |
hypothetical protein JCGZ_20906 [Jatropha curcas] |