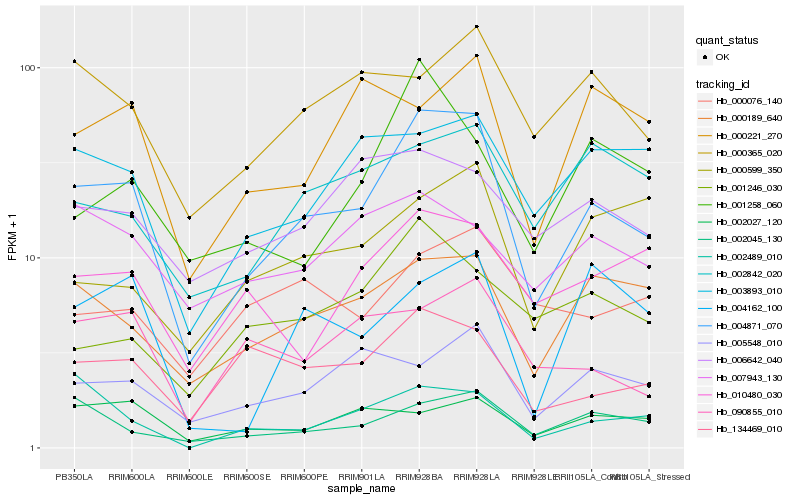
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004871_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632531 [Jatropha curcas] |
| 2 |
Hb_000189_640 |
0.1816235247 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 3 |
Hb_134469_010 |
0.1891759046 |
- |
- |
PREDICTED: protein argonaute 5 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002045_130 |
0.1900351095 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 5 |
Hb_000365_020 |
0.1991511608 |
- |
- |
Vacuolar sorting receptor 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_005548_010 |
0.1995728892 |
- |
- |
PREDICTED: DNA repair protein RAD51 homolog 2 [Jatropha curcas] |
| 7 |
Hb_003893_010 |
0.2006126739 |
- |
- |
hypothetical protein M569_16374 [Genlisea aurea] |
| 8 |
Hb_090855_010 |
0.2011978236 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 9 |
Hb_002842_020 |
0.2034850252 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 10 |
Hb_010480_030 |
0.2051417654 |
- |
- |
PREDICTED: LETM1 and EF-hand domain-containing protein 1, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_006642_040 |
0.2084182918 |
- |
- |
PREDICTED: uncharacterized protein LOC105629489 [Jatropha curcas] |
| 12 |
Hb_004162_100 |
0.2099545409 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000076_140 |
0.2107271914 |
- |
- |
PREDICTED: glucose-6-phosphate/phosphate translocator 1, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_001246_030 |
0.2112621345 |
- |
- |
hypothetical protein JCGZ_14672 [Jatropha curcas] |
| 15 |
Hb_001258_060 |
0.213452635 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 16 |
Hb_002027_120 |
0.215579585 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 17 |
Hb_002489_010 |
0.2156387013 |
- |
- |
hypothetical protein JCGZ_17199 [Jatropha curcas] |
| 18 |
Hb_007943_130 |
0.2170156754 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 19 |
Hb_000221_270 |
0.2174071906 |
- |
- |
hypothetical protein JCGZ_15813 [Jatropha curcas] |
| 20 |
Hb_000599_350 |
0.219414833 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |