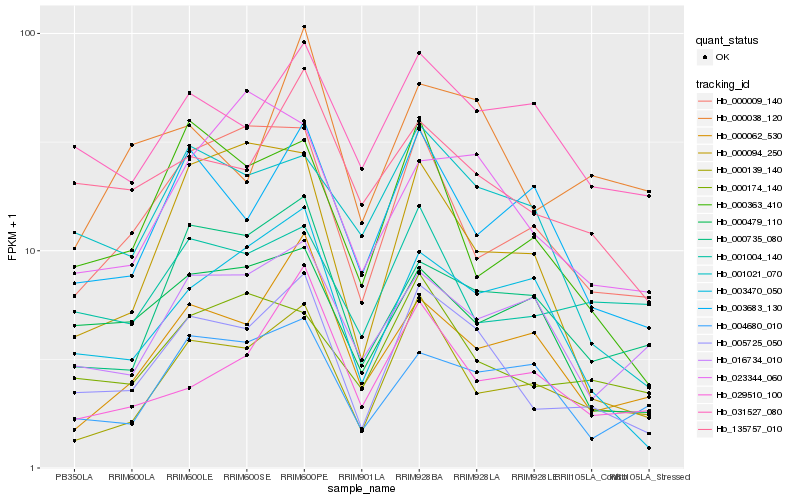
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005725_050 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Jatropha curcas] |
| 2 |
Hb_000139_140 |
0.1549857544 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 3 |
Hb_135757_010 |
0.162565873 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 4 |
Hb_000174_140 |
0.1725672331 |
- |
- |
JHL23J11.8 [Jatropha curcas] |
| 5 |
Hb_000009_140 |
0.1729202767 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 6 |
Hb_000735_080 |
0.1748440486 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 7 |
Hb_001021_070 |
0.1756609188 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_003470_050 |
0.1770896115 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_003683_130 |
0.1771040702 |
- |
- |
PREDICTED: enolase 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000038_120 |
0.1797817488 |
- |
- |
PREDICTED: endoglucanase 9-like [Jatropha curcas] |
| 11 |
Hb_000363_410 |
0.1804601846 |
- |
- |
hypothetical protein JCGZ_00955 [Jatropha curcas] |
| 12 |
Hb_031527_080 |
0.1815779537 |
- |
- |
utp-glucose-1-phosphate uridylyltransferase, putative [Ricinus communis] |
| 13 |
Hb_000062_530 |
0.1852223738 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 14 |
Hb_000479_110 |
0.1880477152 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000094_250 |
0.190101538 |
- |
- |
PREDICTED: uncharacterized protein LOC105633774 isoform X3 [Jatropha curcas] |
| 16 |
Hb_004680_010 |
0.1911378515 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X1 [Jatropha curcas] |
| 17 |
Hb_023344_060 |
0.1928735388 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA11 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001004_140 |
0.1935819018 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 19 |
Hb_029510_100 |
0.1937513557 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 20 |
Hb_016734_010 |
0.1940016568 |
- |
- |
PREDICTED: probable DNA-3-methyladenine glycosylase 2 [Jatropha curcas] |