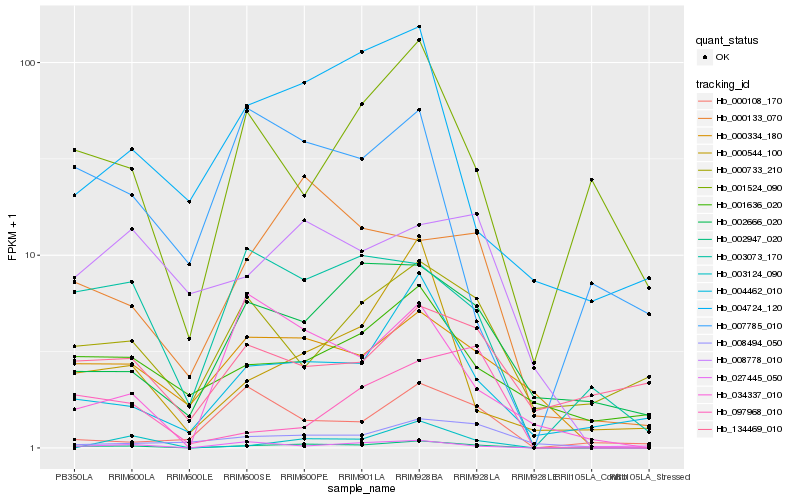
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002947_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103330439 [Prunus mume] |
| 2 |
Hb_000334_180 |
0.2359077414 |
- |
- |
hypothetical protein JCGZ_16779 [Jatropha curcas] |
| 3 |
Hb_004462_010 |
0.2377821104 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 4 |
Hb_000133_070 |
0.2467465293 |
- |
- |
- |
| 5 |
Hb_003073_170 |
0.2525846563 |
- |
- |
- |
| 6 |
Hb_002666_020 |
0.261588945 |
- |
- |
VAMP/SYNAPTOBREVIN-ASSOCIATED protein 27-1 [Populus trichocarpa] |
| 7 |
Hb_034337_010 |
0.2661081895 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 8 |
Hb_008778_010 |
0.2688366412 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_003124_090 |
0.2690946161 |
- |
- |
hypothetical protein CICLE_v10031194mg [Citrus clementina] |
| 10 |
Hb_001636_020 |
0.2759075914 |
- |
- |
Serine/threonine-protein kinase PBS1 [Theobroma cacao] |
| 11 |
Hb_008494_050 |
0.2759232797 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 12 |
Hb_004724_120 |
0.2770197618 |
- |
- |
PREDICTED: ferredoxin, root R-B2 [Jatropha curcas] |
| 13 |
Hb_000544_100 |
0.2772611415 |
- |
- |
hypothetical protein JCGZ_03636 [Jatropha curcas] |
| 14 |
Hb_001524_090 |
0.2815683116 |
- |
- |
PREDICTED: uncharacterized protein LOC105113929 [Populus euphratica] |
| 15 |
Hb_000733_210 |
0.2882692666 |
- |
- |
putative serine/threonine protein phosphatase [Angiostrongylus costaricensis] |
| 16 |
Hb_097968_010 |
0.3017488664 |
- |
- |
hypothetical protein PRUPE_ppa001143mg [Prunus persica] |
| 17 |
Hb_000108_170 |
0.3062229306 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 18 |
Hb_134469_010 |
0.312238563 |
- |
- |
PREDICTED: protein argonaute 5 isoform X1 [Jatropha curcas] |
| 19 |
Hb_027445_050 |
0.315442121 |
- |
- |
PREDICTED: titin isoform X13 [Jatropha curcas] |
| 20 |
Hb_007785_010 |
0.3190865197 |
- |
- |
glutathione peroxidase, putative [Ricinus communis] |