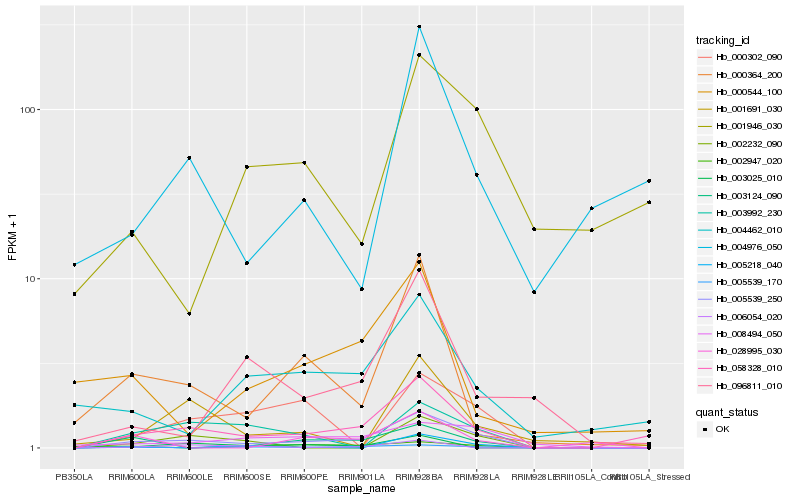
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005539_250 |
0.0 |
- |
- |
- |
| 2 |
Hb_058328_010 |
0.1902308789 |
- |
- |
JHL06B08.2 [Jatropha curcas] |
| 3 |
Hb_003124_090 |
0.2757130202 |
- |
- |
hypothetical protein CICLE_v10031194mg [Citrus clementina] |
| 4 |
Hb_008494_050 |
0.2790572632 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 5 |
Hb_003992_230 |
0.3027942045 |
- |
- |
PREDICTED: uncharacterized protein LOC105634858 [Jatropha curcas] |
| 6 |
Hb_000302_090 |
0.3075010452 |
- |
- |
PREDICTED: protein SGT1 homolog [Prunus mume] |
| 7 |
Hb_000364_200 |
0.3102180775 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_004462_010 |
0.3143798569 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 9 |
Hb_005539_170 |
0.3145232752 |
- |
- |
PREDICTED: uncharacterized protein LOC101502180 [Cicer arietinum] |
| 10 |
Hb_002232_090 |
0.3153143321 |
- |
- |
STS14 protein precursor, putative [Ricinus communis] |
| 11 |
Hb_005218_040 |
0.3251750539 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 8-like [Jatropha curcas] |
| 12 |
Hb_003025_010 |
0.3255522035 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X2 [Populus euphratica] |
| 13 |
Hb_001691_030 |
0.3295033141 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 14 |
Hb_096811_010 |
0.3301874568 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At1g61190 [Vitis vinifera] |
| 15 |
Hb_006054_020 |
0.3395698218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002947_020 |
0.3435985849 |
- |
- |
PREDICTED: uncharacterized protein LOC103330439 [Prunus mume] |
| 17 |
Hb_028995_030 |
0.3436948666 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 18 |
Hb_000544_100 |
0.3446123888 |
- |
- |
hypothetical protein JCGZ_03636 [Jatropha curcas] |
| 19 |
Hb_001946_030 |
0.3463771375 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 20 |
Hb_004976_050 |
0.3467424689 |
- |
- |
- |