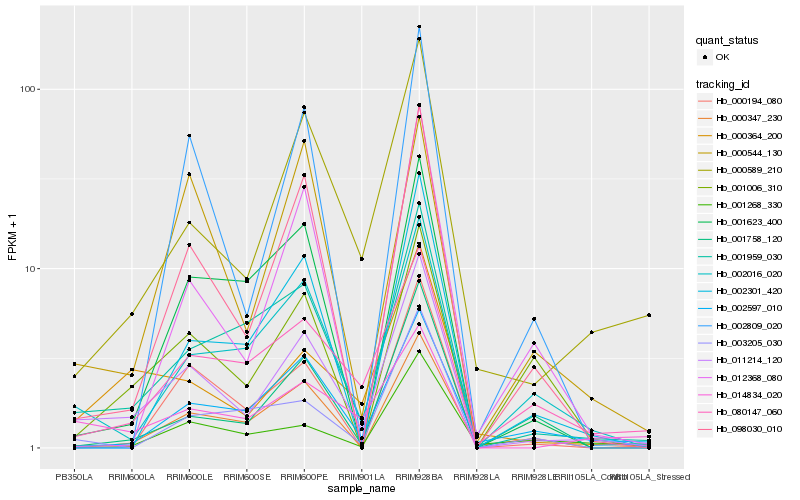
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011214_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636139 [Jatropha curcas] |
| 2 |
Hb_001959_030 |
0.1500314152 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000589_210 |
0.154626142 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |
| 4 |
Hb_098030_010 |
0.1625136509 |
- |
- |
hypothetical protein JCGZ_06251 [Jatropha curcas] |
| 5 |
Hb_000364_200 |
0.1701289925 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_002016_020 |
0.1710165076 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 7 |
Hb_014834_020 |
0.1852715495 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002597_010 |
0.1887399205 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Malus domestica] |
| 9 |
Hb_080147_060 |
0.1913167738 |
- |
- |
PREDICTED: cyclin-dependent kinase F-1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000194_080 |
0.197351265 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 11 |
Hb_000347_230 |
0.1976829822 |
- |
- |
inter-alpha-trypsin inhibitor heavy chain, putative [Ricinus communis] |
| 12 |
Hb_001758_120 |
0.1998582318 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_001006_310 |
0.2028921395 |
- |
- |
PREDICTED: uncharacterized protein LOC105634192 [Jatropha curcas] |
| 14 |
Hb_002809_020 |
0.2051663403 |
- |
- |
PREDICTED: leucoanthocyanidin dioxygenase [Jatropha curcas] |
| 15 |
Hb_001623_400 |
0.2063649723 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002301_420 |
0.2077029383 |
transcription factor |
TF Family: bHLH |
hypothetical protein POPTR_0011s05320g [Populus trichocarpa] |
| 17 |
Hb_000544_130 |
0.2093242214 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 18 |
Hb_012368_080 |
0.2093684218 |
- |
- |
PREDICTED: uncharacterized protein LOC105126277 [Populus euphratica] |
| 19 |
Hb_003205_030 |
0.2095058681 |
- |
- |
Pectinesterase inhibitor, putative [Ricinus communis] |
| 20 |
Hb_001268_330 |
0.2095417619 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |