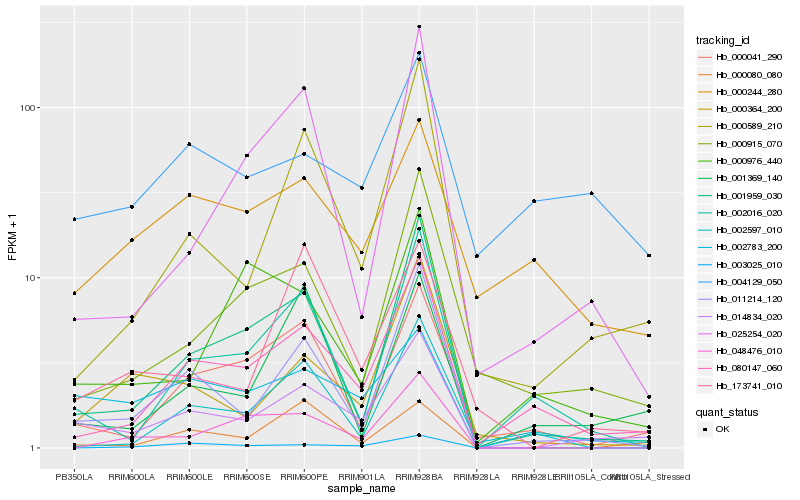
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014834_020 |
0.0 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_011214_120 |
0.1852715495 |
- |
- |
PREDICTED: uncharacterized protein LOC105636139 [Jatropha curcas] |
| 3 |
Hb_080147_060 |
0.1931960612 |
- |
- |
PREDICTED: cyclin-dependent kinase F-1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000589_210 |
0.2017841598 |
- |
- |
PREDICTED: uncharacterized protein LOC105647439 [Jatropha curcas] |
| 5 |
Hb_001959_030 |
0.2068743111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000364_200 |
0.2228583008 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000041_290 |
0.2271198178 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 8 |
Hb_000915_070 |
0.2272637081 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_002783_200 |
0.2374786198 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 10 |
Hb_002016_020 |
0.2391770088 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 11 |
Hb_173741_010 |
0.2445919192 |
- |
- |
PREDICTED: caffeoyl-CoA O-methyltransferase isoform X3 [Musa acuminata subsp. malaccensis] |
| 12 |
Hb_000080_080 |
0.2457025667 |
- |
- |
- |
| 13 |
Hb_002597_010 |
0.2466454657 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Malus domestica] |
| 14 |
Hb_001369_140 |
0.2485099584 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004129_050 |
0.250735723 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 5 [Jatropha curcas] |
| 16 |
Hb_000244_280 |
0.2515795201 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X1 [Jatropha curcas] |
| 17 |
Hb_025254_020 |
0.2532366737 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Vitis vinifera] |
| 18 |
Hb_003025_010 |
0.2544465713 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X2 [Populus euphratica] |
| 19 |
Hb_000976_440 |
0.2655925543 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_048476_010 |
0.2667720627 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Citrus sinensis] |