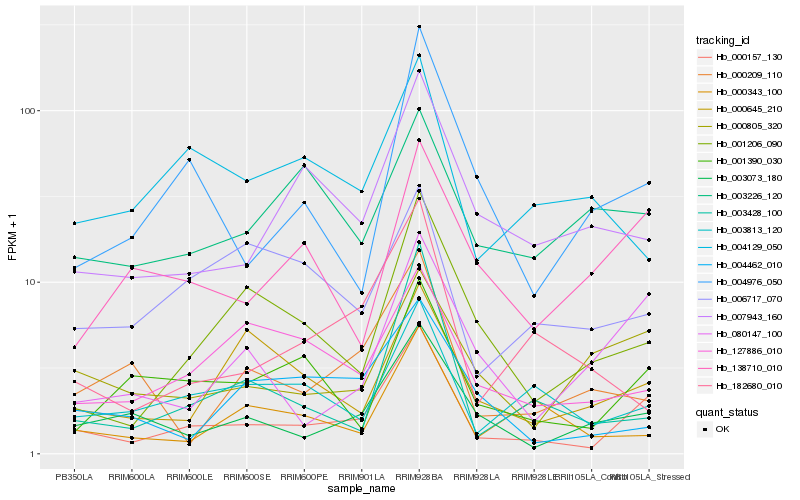
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000157_130 |
0.0 |
- |
- |
PREDICTED: cinnamoyl-CoA reductase 1-like isoform X2 [Populus euphratica] |
| 2 |
Hb_003073_180 |
0.218496723 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 3 |
Hb_080147_100 |
0.2269270703 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN [Jatropha curcas] |
| 4 |
Hb_001390_030 |
0.2294679448 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 5 |
Hb_003813_120 |
0.2322555494 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_000805_320 |
0.2347832809 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Jatropha curcas] |
| 7 |
Hb_004976_050 |
0.2353674874 |
- |
- |
- |
| 8 |
Hb_001206_090 |
0.236295691 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 9 |
Hb_007943_160 |
0.2398918602 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 10 |
Hb_138710_010 |
0.2429038038 |
- |
- |
Adenosylmethionine-8-amino-7-oxononanoate aminotransferase [Gossypium arboreum] |
| 11 |
Hb_003428_100 |
0.2485634832 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 12 |
Hb_000645_210 |
0.2528952632 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_127886_010 |
0.2536931991 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 14 |
Hb_182680_010 |
0.2635112601 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 15 |
Hb_000209_110 |
0.2635650599 |
- |
- |
PREDICTED: receptor-like protein kinase BRI1-like 3 [Jatropha curcas] |
| 16 |
Hb_000343_100 |
0.2655248269 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 17 |
Hb_004462_010 |
0.2683555904 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 18 |
Hb_006717_070 |
0.2690193716 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor CPC-like [Jatropha curcas] |
| 19 |
Hb_003226_120 |
0.2703618522 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004129_050 |
0.2745381444 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 5 [Jatropha curcas] |