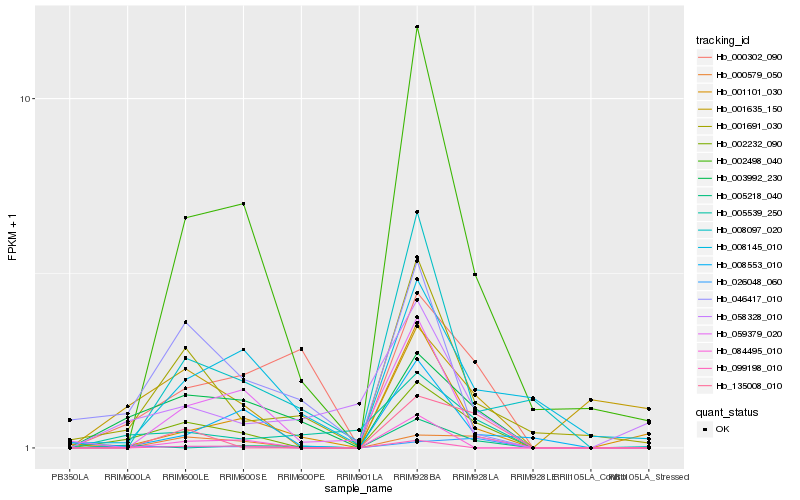
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002232_090 |
0.0 |
- |
- |
STS14 protein precursor, putative [Ricinus communis] |
| 2 |
Hb_003992_230 |
0.2441536349 |
- |
- |
PREDICTED: uncharacterized protein LOC105634858 [Jatropha curcas] |
| 3 |
Hb_002498_040 |
0.267070926 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 4 |
Hb_001691_030 |
0.2879645741 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 5 |
Hb_135008_010 |
0.3017974478 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 6 |
Hb_005539_250 |
0.3153143321 |
- |
- |
- |
| 7 |
Hb_000579_050 |
0.3182880033 |
- |
- |
PREDICTED: peroxidase 9 [Jatropha curcas] |
| 8 |
Hb_026048_060 |
0.319652406 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 9 |
Hb_001101_030 |
0.3309410632 |
- |
- |
hypothetical protein JCGZ_00046 [Jatropha curcas] |
| 10 |
Hb_059379_020 |
0.3321384834 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase 1-like [Populus euphratica] |
| 11 |
Hb_001635_150 |
0.3328569925 |
- |
- |
hypothetical protein JCGZ_24809 [Jatropha curcas] |
| 12 |
Hb_008553_010 |
0.3336236928 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 13 |
Hb_046417_010 |
0.3373594102 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 20 [Jatropha curcas] |
| 14 |
Hb_000302_090 |
0.3380848529 |
- |
- |
PREDICTED: protein SGT1 homolog [Prunus mume] |
| 15 |
Hb_005218_040 |
0.3470615706 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 8-like [Jatropha curcas] |
| 16 |
Hb_008097_020 |
0.3482923841 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 17 |
Hb_008145_010 |
0.3564394454 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 18 |
Hb_099198_010 |
0.3616577983 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At2g25790 isoform X2 [Nicotiana tomentosiformis] |
| 19 |
Hb_058328_010 |
0.3641484861 |
- |
- |
JHL06B08.2 [Jatropha curcas] |
| 20 |
Hb_084495_010 |
0.3653156778 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 [Eucalyptus grandis] |