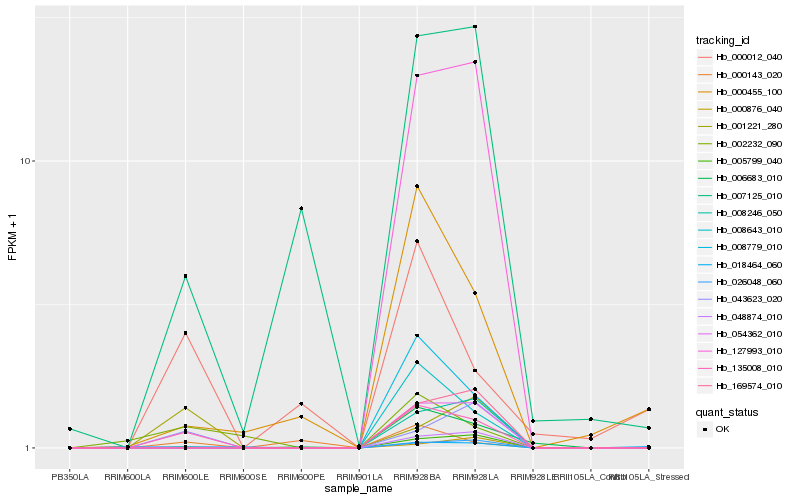
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_135008_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 2 |
Hb_043623_020 |
0.2902501573 |
- |
- |
hypothetical protein JCGZ_17692 [Jatropha curcas] |
| 3 |
Hb_002232_090 |
0.3017974478 |
- |
- |
STS14 protein precursor, putative [Ricinus communis] |
| 4 |
Hb_007125_010 |
0.3021515415 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 5 |
Hb_006683_010 |
0.3112799138 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 6 |
Hb_054362_010 |
0.3151492437 |
- |
- |
PREDICTED: uncharacterized protein LOC105650428 [Jatropha curcas] |
| 7 |
Hb_008779_010 |
0.3169393653 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 8 |
Hb_000012_040 |
0.3206012116 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 9 |
Hb_008643_010 |
0.3207956164 |
- |
- |
hypothetical protein JCGZ_04298 [Jatropha curcas] |
| 10 |
Hb_001221_280 |
0.322141989 |
- |
- |
- |
| 11 |
Hb_127993_010 |
0.3224361511 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X4 [Jatropha curcas] |
| 12 |
Hb_000455_100 |
0.3243541274 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 13 |
Hb_018464_060 |
0.3269906526 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_000876_040 |
0.333146951 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B-like [Jatropha curcas] |
| 15 |
Hb_169574_010 |
0.3414347821 |
- |
- |
PREDICTED: uncharacterized protein LOC104120373 [Nicotiana tomentosiformis] |
| 16 |
Hb_048874_010 |
0.3425930566 |
- |
- |
hypothetical protein POPTR_0016s10870g [Populus trichocarpa] |
| 17 |
Hb_005799_040 |
0.3455030004 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4-like isoform X1 [Gossypium raimondii] |
| 18 |
Hb_008246_050 |
0.3455246857 |
- |
- |
- |
| 19 |
Hb_026048_060 |
0.3457406075 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 20 |
Hb_000143_020 |
0.3465827489 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Jatropha curcas] |