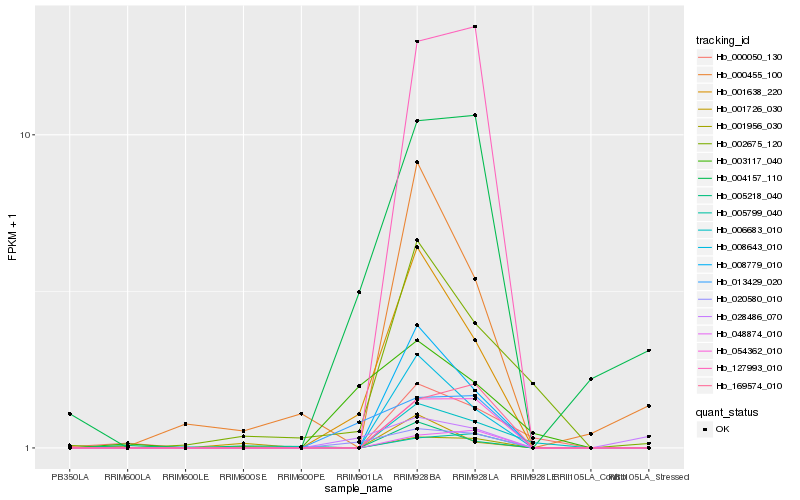
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001638_220 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_008779_010 |
0.172361936 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 3 |
Hb_008643_010 |
0.1727642805 |
- |
- |
hypothetical protein JCGZ_04298 [Jatropha curcas] |
| 4 |
Hb_020580_010 |
0.1833238755 |
- |
- |
- |
| 5 |
Hb_013429_020 |
0.2630728543 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 6 |
Hb_054362_010 |
0.2687503425 |
- |
- |
PREDICTED: uncharacterized protein LOC105650428 [Jatropha curcas] |
| 7 |
Hb_006683_010 |
0.2791001282 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 8 |
Hb_002675_120 |
0.2805966879 |
- |
- |
DNA-repair protein xrcc1, putative [Ricinus communis] |
| 9 |
Hb_000455_100 |
0.2839246168 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 10 |
Hb_127993_010 |
0.2858872245 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X4 [Jatropha curcas] |
| 11 |
Hb_003117_040 |
0.2862207627 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_004157_110 |
0.2939252533 |
- |
- |
- |
| 13 |
Hb_001956_030 |
0.3007066947 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Populus euphratica] |
| 14 |
Hb_000050_130 |
0.3074507579 |
- |
- |
Putative Pol polyprotein from transposon element Bs1 [Triticum urartu] |
| 15 |
Hb_001726_030 |
0.3079614609 |
- |
- |
Ferulic acid 5-hydroxylase 1 [Theobroma cacao] |
| 16 |
Hb_169574_010 |
0.3232693431 |
- |
- |
PREDICTED: uncharacterized protein LOC104120373 [Nicotiana tomentosiformis] |
| 17 |
Hb_048874_010 |
0.3253364072 |
- |
- |
hypothetical protein POPTR_0016s10870g [Populus trichocarpa] |
| 18 |
Hb_005218_040 |
0.3282225878 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 8-like [Jatropha curcas] |
| 19 |
Hb_028486_070 |
0.3293678997 |
- |
- |
hypothetical protein CISIN_1g028119mg [Citrus sinensis] |
| 20 |
Hb_005799_040 |
0.3304515755 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4-like isoform X1 [Gossypium raimondii] |