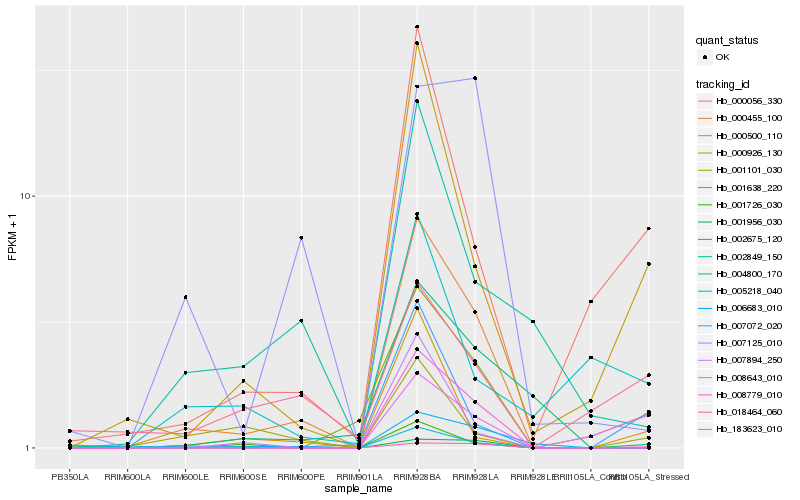
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000455_100 |
0.0 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 2 |
Hb_000926_130 |
0.2286703088 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 3 |
Hb_008643_010 |
0.2304433939 |
- |
- |
hypothetical protein JCGZ_04298 [Jatropha curcas] |
| 4 |
Hb_008779_010 |
0.2305191896 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 5 |
Hb_000056_330 |
0.2369673535 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_002849_150 |
0.2601497669 |
- |
- |
hypothetical protein JCGZ_14367 [Jatropha curcas] |
| 7 |
Hb_006683_010 |
0.2624897733 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 8 |
Hb_005218_040 |
0.2658497998 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate synthase 8-like [Jatropha curcas] |
| 9 |
Hb_018464_060 |
0.2712890423 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_007072_020 |
0.2807465334 |
transcription factor |
TF Family: bHLH |
lMYC5 [Hevea brasiliensis] |
| 11 |
Hb_000500_110 |
0.2827515607 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 12 |
Hb_001638_220 |
0.2839246168 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002675_120 |
0.2888970643 |
- |
- |
DNA-repair protein xrcc1, putative [Ricinus communis] |
| 14 |
Hb_007125_010 |
0.2904213937 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 15 |
Hb_001726_030 |
0.2942849453 |
- |
- |
Ferulic acid 5-hydroxylase 1 [Theobroma cacao] |
| 16 |
Hb_001101_030 |
0.2968981103 |
- |
- |
hypothetical protein JCGZ_00046 [Jatropha curcas] |
| 17 |
Hb_001956_030 |
0.3001830116 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Populus euphratica] |
| 18 |
Hb_007894_250 |
0.3096920991 |
- |
- |
hypothetical protein JCGZ_06575 [Jatropha curcas] |
| 19 |
Hb_004800_170 |
0.3098682926 |
- |
- |
PREDICTED: cytochrome P450 90B1 [Jatropha curcas] |
| 20 |
Hb_183623_010 |
0.310614265 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |