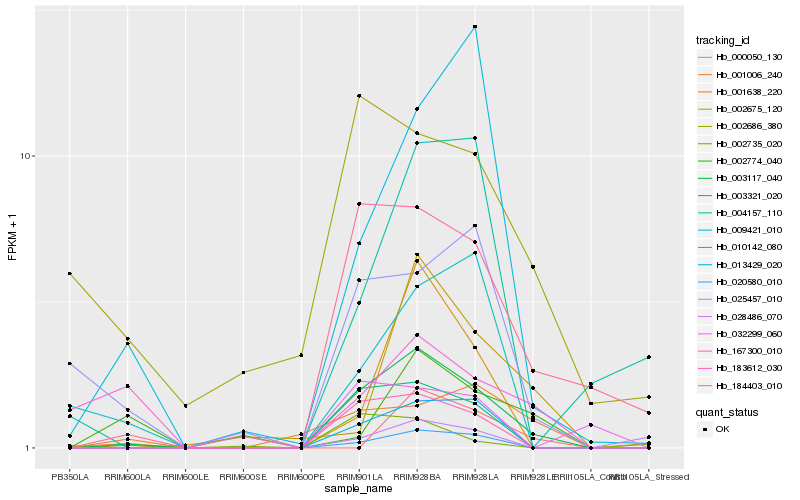
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003117_040 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_003321_020 |
0.2010261471 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Citrus sinensis] |
| 3 |
Hb_020580_010 |
0.2090963255 |
- |
- |
- |
| 4 |
Hb_184403_010 |
0.2119583631 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like [Gossypium raimondii] |
| 5 |
Hb_013429_020 |
0.2222282824 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 6 |
Hb_183612_030 |
0.2691466548 |
- |
- |
PREDICTED: uncharacterized protein LOC105632256 [Jatropha curcas] |
| 7 |
Hb_001638_220 |
0.2862207627 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_025457_010 |
0.2989738978 |
- |
- |
- |
| 9 |
Hb_001006_240 |
0.3133551818 |
- |
- |
- |
| 10 |
Hb_032299_060 |
0.3180984599 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC-like [Elaeis guineensis] |
| 11 |
Hb_009421_010 |
0.3201837632 |
- |
- |
- |
| 12 |
Hb_010142_080 |
0.3262864838 |
- |
- |
PREDICTED: uncharacterized protein LOC105630660 [Jatropha curcas] |
| 13 |
Hb_002686_380 |
0.3293811096 |
- |
- |
- |
| 14 |
Hb_002675_120 |
0.3313640448 |
- |
- |
DNA-repair protein xrcc1, putative [Ricinus communis] |
| 15 |
Hb_004157_110 |
0.3353619975 |
- |
- |
- |
| 16 |
Hb_028486_070 |
0.3386753917 |
- |
- |
hypothetical protein CISIN_1g028119mg [Citrus sinensis] |
| 17 |
Hb_002735_020 |
0.3432344668 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 18 |
Hb_167300_010 |
0.3519288009 |
- |
- |
- |
| 19 |
Hb_002774_040 |
0.3540551166 |
- |
- |
PREDICTED: annexin D1 [Jatropha curcas] |
| 20 |
Hb_000050_130 |
0.3616573538 |
- |
- |
Putative Pol polyprotein from transposon element Bs1 [Triticum urartu] |