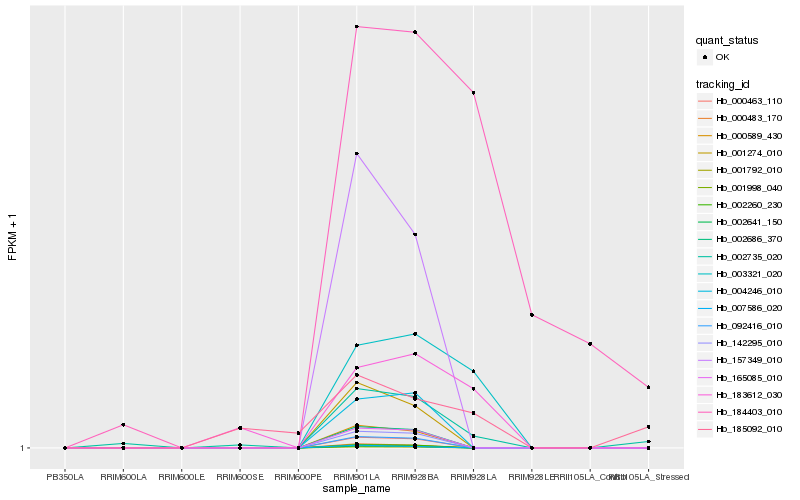
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002735_020 |
0.0 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 2 |
Hb_183612_030 |
0.2831553118 |
- |
- |
PREDICTED: uncharacterized protein LOC105632256 [Jatropha curcas] |
| 3 |
Hb_003321_020 |
0.2876405355 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Citrus sinensis] |
| 4 |
Hb_000463_110 |
0.3041833498 |
- |
- |
- |
| 5 |
Hb_002686_370 |
0.304183981 |
- |
- |
PREDICTED: uncharacterized protein LOC102628436 [Citrus sinensis] |
| 6 |
Hb_092416_010 |
0.3041858541 |
- |
- |
PREDICTED: uncharacterized protein LOC105638802 [Jatropha curcas] |
| 7 |
Hb_002260_230 |
0.3042606299 |
- |
- |
hypothetical protein SORBIDRAFT_05g006180 [Sorghum bicolor] |
| 8 |
Hb_142295_010 |
0.3042678474 |
- |
- |
- |
| 9 |
Hb_001998_040 |
0.3042721269 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_002641_150 |
0.3042986845 |
- |
- |
PREDICTED: stachyose synthase-like [Citrus sinensis] |
| 11 |
Hb_001792_010 |
0.3043196212 |
- |
- |
Uncharacterized protein TCM_043787 [Theobroma cacao] |
| 12 |
Hb_165085_010 |
0.30436898 |
- |
- |
hypothetical protein JCGZ_19461 [Jatropha curcas] |
| 13 |
Hb_000589_430 |
0.305776141 |
- |
- |
hypothetical protein JCGZ_01402 [Jatropha curcas] |
| 14 |
Hb_000483_170 |
0.3072051116 |
transcription factor |
TF Family: NAC |
PREDICTED: protein FEZ [Jatropha curcas] |
| 15 |
Hb_185092_010 |
0.3092403682 |
- |
- |
DNA (cytosine-5)-methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_004246_010 |
0.3102350298 |
- |
- |
- |
| 17 |
Hb_184403_010 |
0.3103495475 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like [Gossypium raimondii] |
| 18 |
Hb_001274_010 |
0.31128297 |
- |
- |
- |
| 19 |
Hb_157349_010 |
0.3131924737 |
- |
- |
PREDICTED: uncharacterized protein LOC102588546 [Solanum tuberosum] |
| 20 |
Hb_007586_020 |
0.3194539576 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |