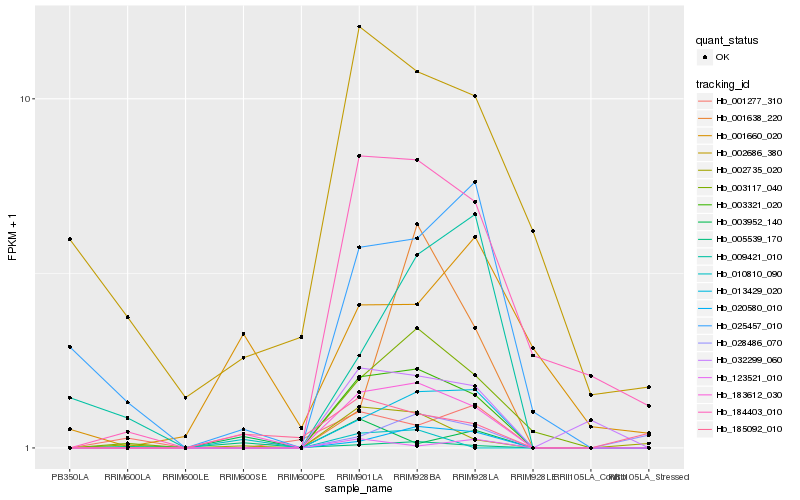
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183612_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632256 [Jatropha curcas] |
| 2 |
Hb_003321_020 |
0.1896366817 |
- |
- |
PREDICTED: TMV resistance protein N-like, partial [Citrus sinensis] |
| 3 |
Hb_013429_020 |
0.2641728061 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 4 |
Hb_003117_040 |
0.2691466548 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_020580_010 |
0.274857487 |
- |
- |
- |
| 6 |
Hb_002735_020 |
0.2831553118 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 7 |
Hb_032299_060 |
0.2993191751 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC-like [Elaeis guineensis] |
| 8 |
Hb_184403_010 |
0.3012954457 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like [Gossypium raimondii] |
| 9 |
Hb_185092_010 |
0.3141779354 |
- |
- |
DNA (cytosine-5)-methyltransferase, putative [Ricinus communis] |
| 10 |
Hb_025457_010 |
0.3407641952 |
- |
- |
- |
| 11 |
Hb_003952_140 |
0.3516593747 |
- |
- |
hypothetical protein POPTR_0001s24960g [Populus trichocarpa] |
| 12 |
Hb_002686_380 |
0.3647035465 |
- |
- |
- |
| 13 |
Hb_005539_170 |
0.3707217586 |
- |
- |
PREDICTED: uncharacterized protein LOC101502180 [Cicer arietinum] |
| 14 |
Hb_010810_090 |
0.371087528 |
- |
- |
- |
| 15 |
Hb_001638_220 |
0.372406348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_123521_010 |
0.3734538272 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 17 |
Hb_028486_070 |
0.3772454462 |
- |
- |
hypothetical protein CISIN_1g028119mg [Citrus sinensis] |
| 18 |
Hb_001660_020 |
0.3820688537 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein Csa_5G648130 [Cucumis sativus] |
| 19 |
Hb_001277_310 |
0.3827853054 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Phoenix dactylifera] |
| 20 |
Hb_009421_010 |
0.3856828486 |
- |
- |
- |