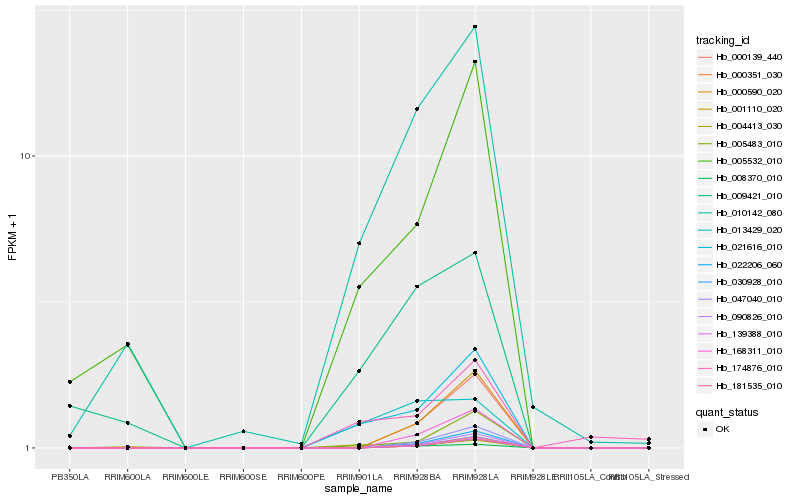
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021616_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_005483_010 |
0.1635994339 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit A [Jatropha curcas] |
| 3 |
Hb_010142_080 |
0.180693429 |
- |
- |
PREDICTED: uncharacterized protein LOC105630660 [Jatropha curcas] |
| 4 |
Hb_005532_010 |
0.1900370029 |
- |
- |
- |
| 5 |
Hb_174876_010 |
0.2260801136 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 6 |
Hb_013429_020 |
0.2276286925 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 7 |
Hb_008370_010 |
0.2346528157 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_004413_030 |
0.2377045077 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 9 |
Hb_168311_010 |
0.2499714306 |
- |
- |
- |
| 10 |
Hb_000139_440 |
0.2503056223 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_000590_020 |
0.2511437998 |
- |
- |
hypothetical protein JCGZ_18748 [Jatropha curcas] |
| 12 |
Hb_047040_010 |
0.2516361918 |
- |
- |
integrase [Gossypium herbaceum] |
| 13 |
Hb_022206_060 |
0.2527130506 |
- |
- |
- |
| 14 |
Hb_090826_010 |
0.2533820445 |
- |
- |
PREDICTED: uncharacterized protein LOC105786910 [Gossypium raimondii] |
| 15 |
Hb_030928_010 |
0.2541828655 |
- |
- |
- |
| 16 |
Hb_181535_010 |
0.2543076165 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 17 |
Hb_000351_030 |
0.2547452429 |
- |
- |
putative NBS-LRR type disease resistance protein [Prunus persica] |
| 18 |
Hb_139388_010 |
0.2548050721 |
- |
- |
PREDICTED: uncharacterized protein LOC104222982, partial [Nicotiana sylvestris] |
| 19 |
Hb_009421_010 |
0.254860165 |
- |
- |
- |
| 20 |
Hb_001110_020 |
0.2549060198 |
- |
- |
hypothetical protein JCGZ_03693 [Jatropha curcas] |