| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
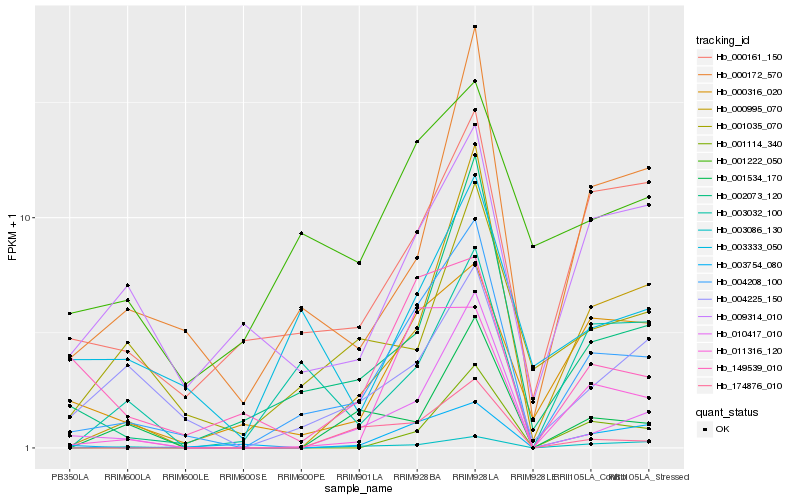
Hb_004208_100 |
0.0 |
- |
- |
Ribosomal protein L3 plastid isoform 1 [Theobroma cacao] |
| 2 |
Hb_002073_120 |
0.1943669694 |
- |
- |
Adenine nucleotide alpha hydrolases-like superfamily protein [Theobroma cacao] |
| 3 |
Hb_000172_570 |
0.2142737631 |
- |
- |
PREDICTED: uncharacterized protein LOC105636518 isoform X1 [Jatropha curcas] |
| 4 |
Hb_010417_010 |
0.2184727501 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000995_070 |
0.2200745013 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 6 |
Hb_011316_120 |
0.2319027862 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH118-like [Jatropha curcas] |
| 7 |
Hb_003754_080 |
0.2355637799 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 8 |
Hb_000161_150 |
0.2368764225 |
- |
- |
PREDICTED: protein argonaute 2-like [Jatropha curcas] |
| 9 |
Hb_004225_150 |
0.2396775345 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger protein 36, C3H1 type-like 3 [Jatropha curcas] |
| 10 |
Hb_000316_020 |
0.2455031429 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_003086_130 |
0.2514337359 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 12 |
Hb_009314_010 |
0.2520687469 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 13 |
Hb_174876_010 |
0.2555264931 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 14 |
Hb_001222_050 |
0.2580381486 |
- |
- |
hypothetical protein CICLE_v100161911mg, partial [Citrus clementina] |
| 15 |
Hb_003032_100 |
0.2583874691 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 16 |
Hb_149539_010 |
0.2591913679 |
- |
- |
- |
| 17 |
Hb_003333_050 |
0.2608938463 |
- |
- |
hypothetical protein JCGZ_20906 [Jatropha curcas] |
| 18 |
Hb_001035_070 |
0.2636243848 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.7 [Jatropha curcas] |
| 19 |
Hb_001114_340 |
0.2637825557 |
- |
- |
hypothetical protein JCGZ_13805 [Jatropha curcas] |
| 20 |
Hb_001534_170 |
0.2646040398 |
- |
- |
Uncharacterized protein TCM_038456 [Theobroma cacao] |