| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
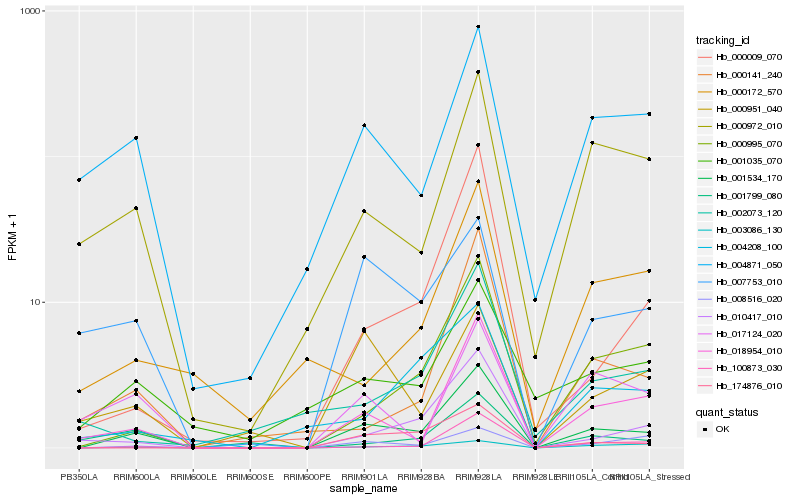
Hb_001534_170 |
0.0 |
- |
- |
Uncharacterized protein TCM_038456 [Theobroma cacao] |
| 2 |
Hb_018954_010 |
0.1941763319 |
- |
- |
- |
| 3 |
Hb_000995_070 |
0.2192738033 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 4 |
Hb_010417_010 |
0.220174143 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 5 |
Hb_100873_030 |
0.2204437373 |
- |
- |
PREDICTED: uncharacterized protein LOC105795653 [Gossypium raimondii] |
| 6 |
Hb_004871_050 |
0.2249791744 |
- |
- |
hypothetical protein F383_22684 [Gossypium arboreum] |
| 7 |
Hb_174876_010 |
0.2306329224 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 8 |
Hb_008516_020 |
0.2386071739 |
- |
- |
PREDICTED: putative B3 domain-containing protein At2g27410 [Jatropha curcas] |
| 9 |
Hb_000972_010 |
0.2389331236 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 10 |
Hb_007753_010 |
0.2498472104 |
- |
- |
- |
| 11 |
Hb_000141_240 |
0.2502054694 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000009_070 |
0.2508160383 |
- |
- |
PREDICTED: globulin-1 S allele [Jatropha curcas] |
| 13 |
Hb_003086_130 |
0.2530051051 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 14 |
Hb_017124_020 |
0.2577845718 |
- |
- |
hypothetical protein PRUPE_ppa015539mg [Prunus persica] |
| 15 |
Hb_001035_070 |
0.2588094645 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.7 [Jatropha curcas] |
| 16 |
Hb_004208_100 |
0.2646040398 |
- |
- |
Ribosomal protein L3 plastid isoform 1 [Theobroma cacao] |
| 17 |
Hb_000951_040 |
0.2651635158 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_09931 [Jatropha curcas] |
| 18 |
Hb_002073_120 |
0.2660518418 |
- |
- |
Adenine nucleotide alpha hydrolases-like superfamily protein [Theobroma cacao] |
| 19 |
Hb_001799_080 |
0.2700889692 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 20 |
Hb_000172_570 |
0.281497364 |
- |
- |
PREDICTED: uncharacterized protein LOC105636518 isoform X1 [Jatropha curcas] |