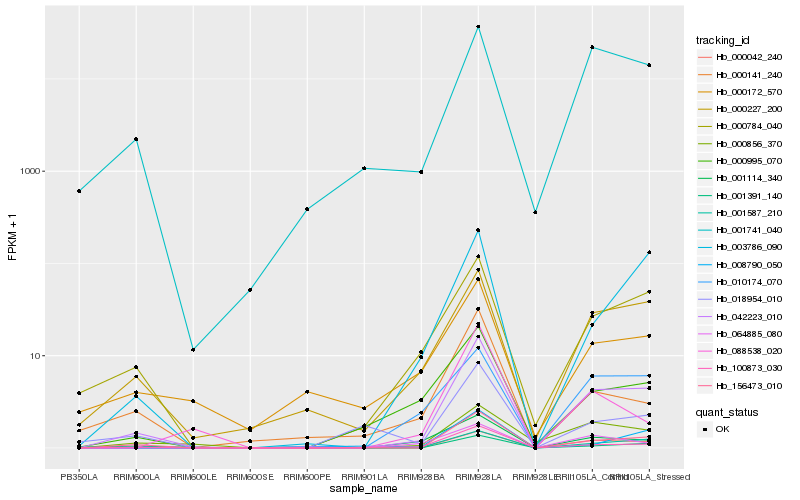
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_042223_010 |
0.0 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 2 |
Hb_156473_010 |
0.1110086831 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 3 |
Hb_100873_030 |
0.1547433089 |
- |
- |
PREDICTED: uncharacterized protein LOC105795653 [Gossypium raimondii] |
| 4 |
Hb_000042_240 |
0.1628015702 |
- |
- |
- |
| 5 |
Hb_000227_200 |
0.172919498 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 3 [Populus euphratica] |
| 6 |
Hb_001587_210 |
0.1782198072 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 [Jatropha curcas] |
| 7 |
Hb_000784_040 |
0.1796633504 |
- |
- |
hypothetical protein POPTR_0013s11550g [Populus trichocarpa] |
| 8 |
Hb_000995_070 |
0.201039686 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 9 |
Hb_001114_340 |
0.2063237452 |
- |
- |
hypothetical protein JCGZ_13805 [Jatropha curcas] |
| 10 |
Hb_001741_040 |
0.2105771578 |
rubber biosynthesis |
Gene Name: REF3 |
REF-like stress related protein 1 [Hevea brasiliensis] |
| 11 |
Hb_010174_070 |
0.2183291598 |
- |
- |
- |
| 12 |
Hb_003786_090 |
0.2210057199 |
- |
- |
hypothetical protein JCGZ_19199 [Jatropha curcas] |
| 13 |
Hb_000856_370 |
0.2224806437 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 14 |
Hb_000141_240 |
0.2247841787 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000172_570 |
0.2268520879 |
- |
- |
PREDICTED: uncharacterized protein LOC105636518 isoform X1 [Jatropha curcas] |
| 16 |
Hb_018954_010 |
0.2278277675 |
- |
- |
- |
| 17 |
Hb_001391_140 |
0.2352580295 |
- |
- |
PREDICTED: uncharacterized protein LOC101293660 [Fragaria vesca subsp. vesca] |
| 18 |
Hb_064885_080 |
0.2355719565 |
- |
- |
- |
| 19 |
Hb_088538_020 |
0.2384332207 |
- |
- |
hypothetical protein JCGZ_12178 [Jatropha curcas] |
| 20 |
Hb_008790_050 |
0.242310336 |
- |
- |
conserved hypothetical protein [Ricinus communis] |