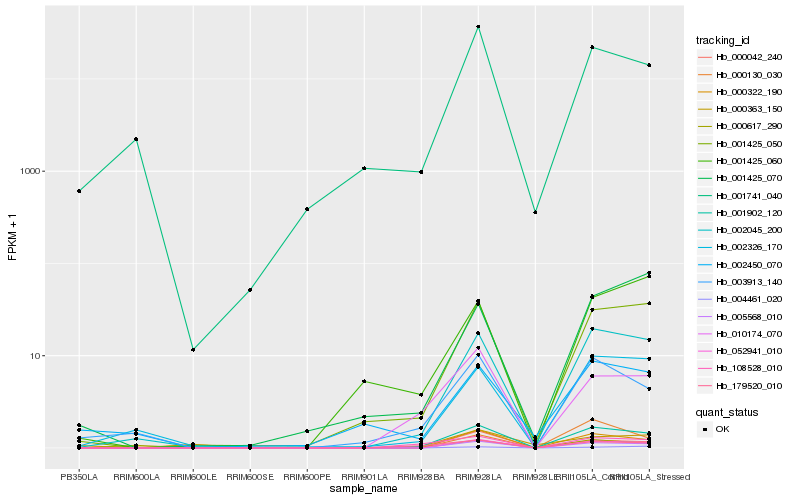
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_052941_010 |
0.0 |
- |
- |
PREDICTED: probable peroxidase 61 [Jatropha curcas] |
| 2 |
Hb_005568_010 |
0.008503733 |
- |
- |
- |
| 3 |
Hb_001902_120 |
0.1023938594 |
- |
- |
PREDICTED: uncharacterized protein LOC105648455 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002045_200 |
0.1125742922 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001425_050 |
0.1360622119 |
- |
- |
- |
| 6 |
Hb_000042_240 |
0.1415486549 |
- |
- |
- |
| 7 |
Hb_002326_170 |
0.186241926 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 8 |
Hb_004461_020 |
0.2041563589 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 9 |
Hb_010174_070 |
0.2047333353 |
- |
- |
- |
| 10 |
Hb_001741_040 |
0.208898892 |
rubber biosynthesis |
Gene Name: REF3 |
REF-like stress related protein 1 [Hevea brasiliensis] |
| 11 |
Hb_003913_140 |
0.2132843996 |
- |
- |
PREDICTED: uncharacterized protein LOC105640324 [Jatropha curcas] |
| 12 |
Hb_179520_010 |
0.2248034966 |
- |
- |
PREDICTED: uncharacterized protein LOC105763691 [Gossypium raimondii] |
| 13 |
Hb_108528_010 |
0.2367206765 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_000130_030 |
0.242266882 |
- |
- |
PREDICTED: small ubiquitin-related modifier 2-A-like [Eucalyptus grandis] |
| 15 |
Hb_000617_290 |
0.2437867228 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002450_070 |
0.2464313623 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 17 |
Hb_001425_060 |
0.24892871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000363_150 |
0.2497872016 |
- |
- |
hypothetical protein POPTR_0011s09040g [Populus trichocarpa] |
| 19 |
Hb_001425_070 |
0.2511192642 |
- |
- |
- |
| 20 |
Hb_000322_190 |
0.2535756013 |
- |
- |
- |