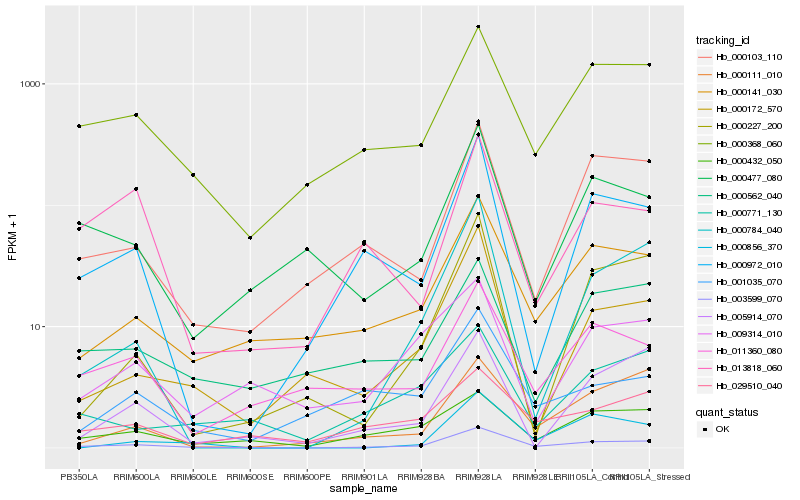
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003599_070 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_000141_030 |
0.1653941357 |
- |
- |
PREDICTED: scarecrow-like protein 22 [Jatropha curcas] |
| 3 |
Hb_000368_060 |
0.1663444173 |
- |
- |
thioredoxin H-type 6 [Hevea brasiliensis] |
| 4 |
Hb_000972_010 |
0.1775849173 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 5 |
Hb_000103_110 |
0.188930703 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000784_040 |
0.1901434403 |
- |
- |
hypothetical protein POPTR_0013s11550g [Populus trichocarpa] |
| 7 |
Hb_000172_570 |
0.1928354427 |
- |
- |
PREDICTED: uncharacterized protein LOC105636518 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000432_050 |
0.1929552701 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |
| 9 |
Hb_000477_080 |
0.1930677847 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11 [Jatropha curcas] |
| 10 |
Hb_011360_080 |
0.1955155032 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_029510_040 |
0.1964359963 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g23330 [Vitis vinifera] |
| 12 |
Hb_001035_070 |
0.2074860062 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.7 [Jatropha curcas] |
| 13 |
Hb_000111_010 |
0.2087147814 |
- |
- |
DNA-directed RNA polymerase, putative [Ricinus communis] |
| 14 |
Hb_005914_070 |
0.2114848843 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000227_200 |
0.2117151228 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 3 [Populus euphratica] |
| 16 |
Hb_013818_060 |
0.2118600273 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 17 |
Hb_009314_010 |
0.2120074384 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 18 |
Hb_000771_130 |
0.2125268781 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 19 |
Hb_000562_040 |
0.2167436129 |
- |
- |
PREDICTED: auxin response factor 5 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000856_370 |
0.21693094 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |