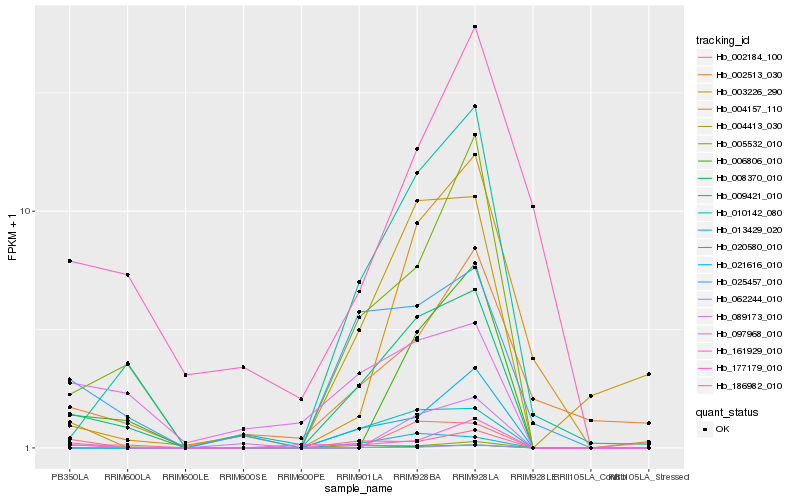
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009421_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_008370_010 |
0.152581588 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_010142_080 |
0.1802401648 |
- |
- |
PREDICTED: uncharacterized protein LOC105630660 [Jatropha curcas] |
| 4 |
Hb_005532_010 |
0.2010122488 |
- |
- |
- |
| 5 |
Hb_025457_010 |
0.2036683764 |
- |
- |
- |
| 6 |
Hb_013429_020 |
0.2239832233 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 7 |
Hb_020580_010 |
0.2416357286 |
- |
- |
- |
| 8 |
Hb_004157_110 |
0.2508878621 |
- |
- |
- |
| 9 |
Hb_021616_010 |
0.254860165 |
- |
- |
- |
| 10 |
Hb_006806_010 |
0.2583591253 |
- |
- |
- |
| 11 |
Hb_004413_030 |
0.2618454621 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 12 |
Hb_097968_010 |
0.2768066555 |
- |
- |
hypothetical protein PRUPE_ppa001143mg [Prunus persica] |
| 13 |
Hb_003226_290 |
0.2970482331 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_002513_030 |
0.2972711174 |
- |
- |
PREDICTED: alpha-L-fucosidase 2-like isoform X2 [Elaeis guineensis] |
| 15 |
Hb_161929_010 |
0.2995614277 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 16 |
Hb_062244_010 |
0.300540558 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_002184_100 |
0.3006770974 |
- |
- |
hypothetical protein PRUPE_ppa020120mg [Prunus persica] |
| 18 |
Hb_089173_010 |
0.3022417875 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 19 |
Hb_186982_010 |
0.3134469055 |
- |
- |
PREDICTED: uncharacterized protein LOC104120222 [Nicotiana tomentosiformis] |
| 20 |
Hb_177179_010 |
0.3150217037 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Populus euphratica] |