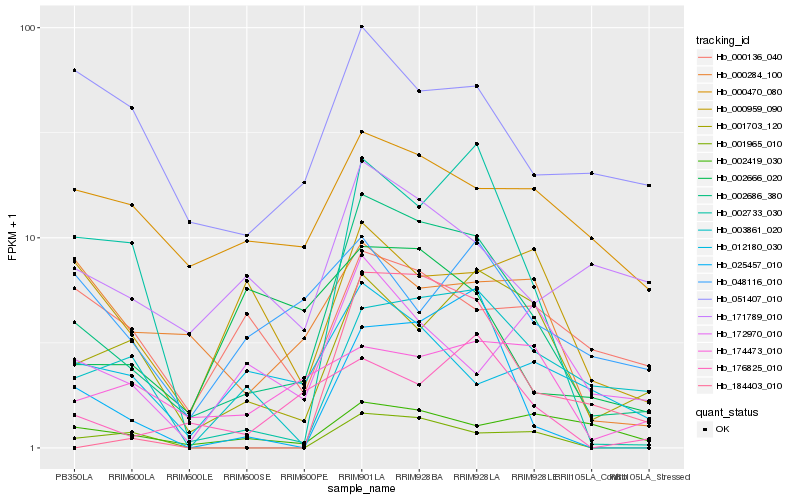
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002686_380 |
0.0 |
- |
- |
- |
| 2 |
Hb_001703_120 |
0.241186042 |
- |
- |
PREDICTED: uncharacterized protein LOC105116966 isoform X2 [Populus euphratica] |
| 3 |
Hb_002733_030 |
0.2481382205 |
- |
- |
PREDICTED: uncharacterized protein LOC104248159 [Nicotiana sylvestris] |
| 4 |
Hb_003861_020 |
0.2567620754 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like isoform X2 [Nelumbo nucifera] |
| 5 |
Hb_025457_010 |
0.2568406975 |
- |
- |
- |
| 6 |
Hb_176825_010 |
0.2595224898 |
- |
- |
PREDICTED: uncharacterized protein LOC105630857 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001965_010 |
0.2682675983 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 8 |
Hb_171789_010 |
0.2743297332 |
- |
- |
phosphoenolpyruvate carboxylase [Glycine max] |
| 9 |
Hb_184403_010 |
0.2766761057 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like [Gossypium raimondii] |
| 10 |
Hb_012180_030 |
0.2779138312 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_051407_010 |
0.2779648078 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 12 |
Hb_002419_030 |
0.2823069655 |
- |
- |
- |
| 13 |
Hb_002666_020 |
0.2850823139 |
- |
- |
VAMP/SYNAPTOBREVIN-ASSOCIATED protein 27-1 [Populus trichocarpa] |
| 14 |
Hb_174473_010 |
0.2864275317 |
- |
- |
PREDICTED: uncharacterized protein LOC105775505 [Gossypium raimondii] |
| 15 |
Hb_000284_100 |
0.2872635998 |
- |
- |
PREDICTED: transcription factor GTE11 isoform X2 [Populus euphratica] |
| 16 |
Hb_000470_080 |
0.2880858113 |
- |
- |
Caldesmon, putative [Ricinus communis] |
| 17 |
Hb_172970_010 |
0.289082136 |
- |
- |
PREDICTED: uncharacterized protein LOC105637608 [Jatropha curcas] |
| 18 |
Hb_000136_040 |
0.2903366141 |
- |
- |
PREDICTED: protein RRP5 homolog [Jatropha curcas] |
| 19 |
Hb_000959_090 |
0.2917777718 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_048116_010 |
0.29375241 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |