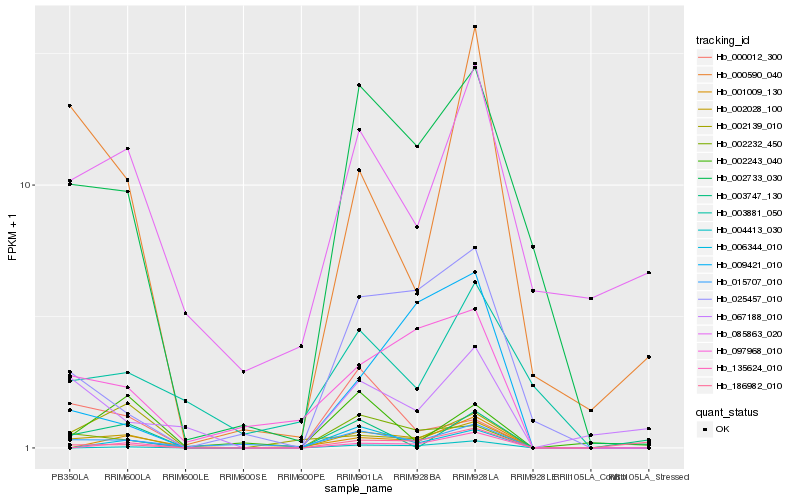
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002028_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC102624085 [Citrus sinensis] |
| 2 |
Hb_002733_030 |
0.2060206707 |
- |
- |
PREDICTED: uncharacterized protein LOC104248159 [Nicotiana sylvestris] |
| 3 |
Hb_002232_450 |
0.2064400373 |
- |
- |
PREDICTED: probable methyltransferase PMT11 [Jatropha curcas] |
| 4 |
Hb_015707_010 |
0.2275796525 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 5 |
Hb_097968_010 |
0.2538952285 |
- |
- |
hypothetical protein PRUPE_ppa001143mg [Prunus persica] |
| 6 |
Hb_000590_040 |
0.2622456144 |
- |
- |
hypothetical protein MPER_09217 [Moniliophthora perniciosa FA553] |
| 7 |
Hb_025457_010 |
0.2716243965 |
- |
- |
- |
| 8 |
Hb_135624_010 |
0.2719802516 |
- |
- |
PREDICTED: inositol transporter 4-like [Jatropha curcas] |
| 9 |
Hb_004413_030 |
0.2897840764 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 10 |
Hb_006344_010 |
0.291936817 |
- |
- |
- |
| 11 |
Hb_002139_010 |
0.2969177176 |
- |
- |
- |
| 12 |
Hb_186982_010 |
0.298417117 |
- |
- |
PREDICTED: uncharacterized protein LOC104120222 [Nicotiana tomentosiformis] |
| 13 |
Hb_001009_130 |
0.2999437986 |
- |
- |
hypothetical protein JCGZ_11336 [Jatropha curcas] |
| 14 |
Hb_085863_020 |
0.3014914 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 15 |
Hb_067188_010 |
0.3036632156 |
- |
- |
- |
| 16 |
Hb_000012_300 |
0.3061829667 |
- |
- |
- |
| 17 |
Hb_003747_130 |
0.3177154702 |
- |
- |
- |
| 18 |
Hb_002243_040 |
0.3182705023 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00227 [Jatropha curcas] |
| 19 |
Hb_003881_050 |
0.3200325961 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 20 |
Hb_009421_010 |
0.3204814932 |
- |
- |
- |