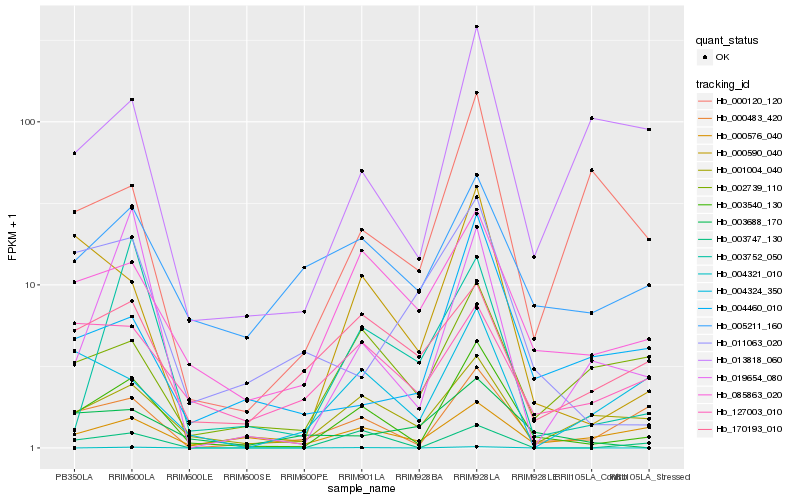
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003540_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_000590_040 |
0.2324418288 |
- |
- |
hypothetical protein MPER_09217 [Moniliophthora perniciosa FA553] |
| 3 |
Hb_000483_420 |
0.2447748647 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 4 [Jatropha curcas] |
| 4 |
Hb_001004_040 |
0.2503605331 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_085863_020 |
0.2527982409 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 6 |
Hb_003747_130 |
0.2557512648 |
- |
- |
- |
| 7 |
Hb_019654_080 |
0.2772392106 |
- |
- |
- |
| 8 |
Hb_002739_110 |
0.2787460277 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 9 |
Hb_013818_060 |
0.2828684069 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 10 |
Hb_004460_010 |
0.2839193733 |
- |
- |
PREDICTED: SKI/DACH domain-containing protein 1-like [Jatropha curcas] |
| 11 |
Hb_003752_050 |
0.2953749901 |
- |
- |
hypothetical protein JCGZ_15078 [Jatropha curcas] |
| 12 |
Hb_000576_040 |
0.2959236162 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g15130 [Jatropha curcas] |
| 13 |
Hb_004321_010 |
0.2976188059 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_004324_350 |
0.2979873884 |
- |
- |
PREDICTED: uncharacterized protein LOC103484146 [Cucumis melo] |
| 15 |
Hb_005211_160 |
0.2997347079 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 16 |
Hb_127003_010 |
0.3024251013 |
- |
- |
PREDICTED: uncharacterized protein LOC105633613 [Jatropha curcas] |
| 17 |
Hb_000120_120 |
0.305199367 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 2 [Jatropha curcas] |
| 18 |
Hb_003688_170 |
0.3061201696 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 isoform X2 [Cucumis sativus] |
| 19 |
Hb_011063_020 |
0.306899024 |
- |
- |
PREDICTED: ribosomal L1 domain-containing protein 1-like [Jatropha curcas] |
| 20 |
Hb_170193_010 |
0.3069123842 |
- |
- |
JHL25H03.10 [Jatropha curcas] |