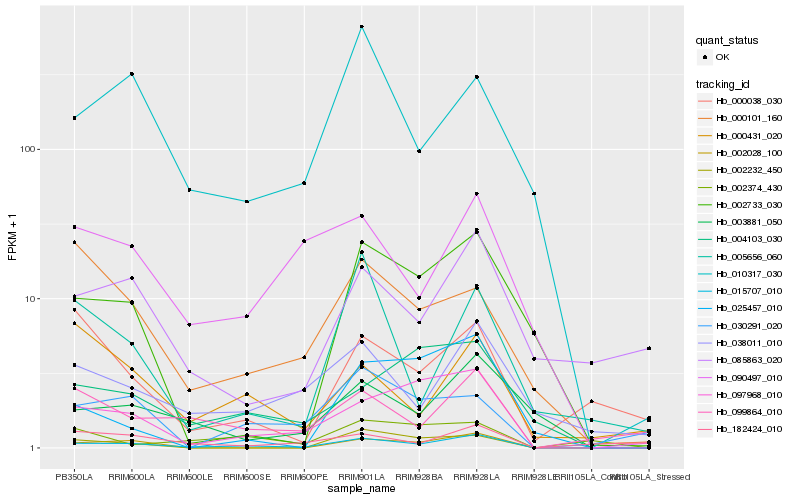
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015707_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 2 |
Hb_097968_010 |
0.2096976337 |
- |
- |
hypothetical protein PRUPE_ppa001143mg [Prunus persica] |
| 3 |
Hb_002028_100 |
0.2275796525 |
- |
- |
PREDICTED: uncharacterized protein LOC102624085 [Citrus sinensis] |
| 4 |
Hb_003881_050 |
0.2395173954 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 5 |
Hb_038011_010 |
0.2416960949 |
- |
- |
hypothetical protein POPTR_0031s004401g, partial [Populus trichocarpa] |
| 6 |
Hb_099864_010 |
0.2476172562 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 7 |
Hb_090497_010 |
0.2477512545 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 8 |
Hb_010317_030 |
0.2524451714 |
- |
- |
unknown [Picea sitchensis] |
| 9 |
Hb_002733_030 |
0.2625360167 |
- |
- |
PREDICTED: uncharacterized protein LOC104248159 [Nicotiana sylvestris] |
| 10 |
Hb_004103_030 |
0.2670467363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_085863_020 |
0.2672972636 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 1 [Jatropha curcas] |
| 12 |
Hb_030291_020 |
0.2729317889 |
- |
- |
PREDICTED: protein argonaute 5 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002232_450 |
0.2733401577 |
- |
- |
PREDICTED: probable methyltransferase PMT11 [Jatropha curcas] |
| 14 |
Hb_005656_060 |
0.2797749355 |
- |
- |
- |
| 15 |
Hb_000431_020 |
0.2830520519 |
- |
- |
hypothetical protein JCGZ_06801 [Jatropha curcas] |
| 16 |
Hb_000101_160 |
0.2859993251 |
- |
- |
PREDICTED: uncharacterized protein LOC105133607 isoform X1 [Populus euphratica] |
| 17 |
Hb_182424_010 |
0.2886382565 |
- |
- |
PREDICTED: U-box domain-containing protein 51 [Jatropha curcas] |
| 18 |
Hb_002374_430 |
0.289258456 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_025457_010 |
0.293449204 |
- |
- |
- |
| 20 |
Hb_000038_030 |
0.2940109912 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7-like [Glycine max] |