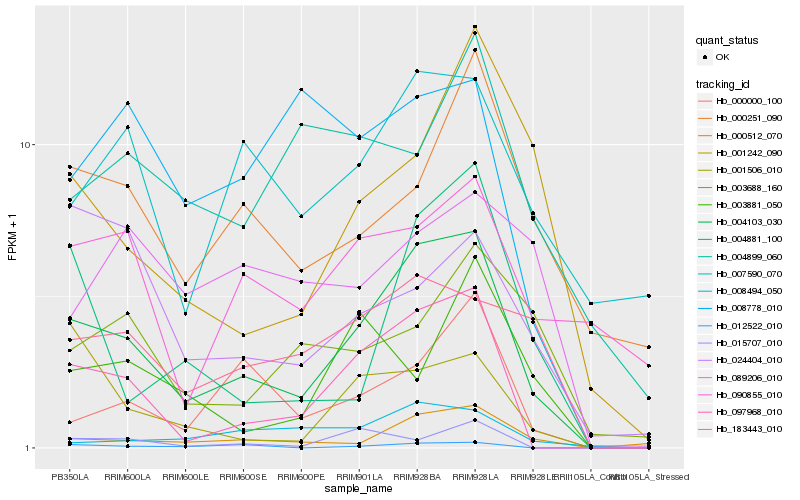
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004103_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_097968_010 |
0.1688877857 |
- |
- |
hypothetical protein PRUPE_ppa001143mg [Prunus persica] |
| 3 |
Hb_183443_010 |
0.2134396077 |
- |
- |
PREDICTED: uncharacterized protein LOC105786773 [Gossypium raimondii] |
| 4 |
Hb_000512_070 |
0.2149288207 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71250-like [Jatropha curcas] |
| 5 |
Hb_001242_090 |
0.2372058322 |
- |
- |
40S ribosomal protein S18 [Colletotrichum fioriniae PJ7] |
| 6 |
Hb_000000_100 |
0.2383455025 |
- |
- |
hypothetical protein POPTR_0002s13260g [Populus trichocarpa] |
| 7 |
Hb_007590_070 |
0.2414018408 |
- |
- |
PREDICTED: inositol 3-kinase [Jatropha curcas] |
| 8 |
Hb_004881_100 |
0.2427183357 |
- |
- |
Phosphoribosyltransferase family protein isoform 4 [Theobroma cacao] |
| 9 |
Hb_000251_090 |
0.2463040204 |
- |
- |
PREDICTED: two-pore potassium channel 3-like [Jatropha curcas] |
| 10 |
Hb_008494_050 |
0.2468281323 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 11 |
Hb_012522_010 |
0.2500981247 |
- |
- |
hypothetical protein VITISV_003261 [Vitis vinifera] |
| 12 |
Hb_001506_010 |
0.2534161709 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 13 |
Hb_090855_010 |
0.2632694984 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 14 |
Hb_003688_160 |
0.26336963 |
- |
- |
Presenilin-like family protein [Populus trichocarpa] |
| 15 |
Hb_008778_010 |
0.2644629261 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_024404_010 |
0.2646387915 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Vitis vinifera] |
| 17 |
Hb_003881_050 |
0.2648294216 |
- |
- |
PREDICTED: uncharacterized protein LOC104213140 [Nicotiana sylvestris] |
| 18 |
Hb_015707_010 |
0.2670467363 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 19 |
Hb_004899_060 |
0.2700676534 |
- |
- |
46 kDa ketoavyl-ACP synthase [Ricinus communis] |
| 20 |
Hb_089206_010 |
0.2728756764 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |