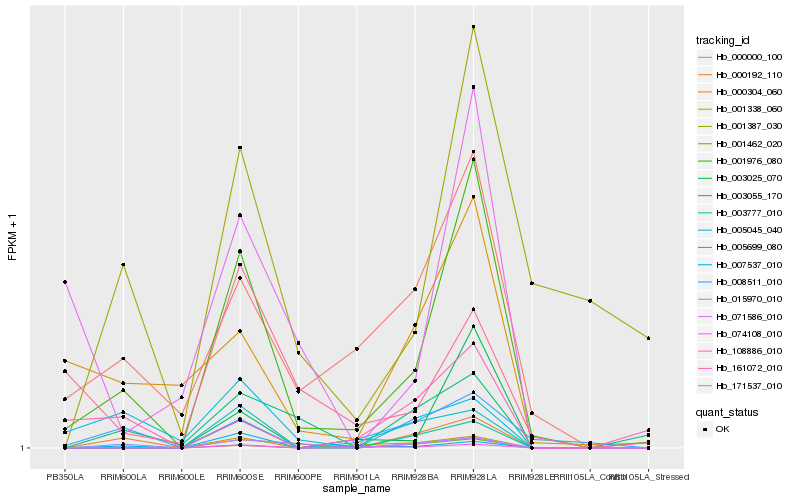
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001976_080 |
0.0 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor AIL6 [Jatropha curcas] |
| 2 |
Hb_000000_100 |
0.2501954596 |
- |
- |
hypothetical protein POPTR_0002s13260g [Populus trichocarpa] |
| 3 |
Hb_008511_010 |
0.2778911401 |
- |
- |
PREDICTED: putative proline-rich receptor-like protein kinase PERK6 [Jatropha curcas] |
| 4 |
Hb_000304_060 |
0.2953828997 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0010s15360g [Populus trichocarpa] |
| 5 |
Hb_000192_110 |
0.2971223505 |
- |
- |
PREDICTED: uncharacterized protein LOC105646856 [Jatropha curcas] |
| 6 |
Hb_015970_010 |
0.2992477271 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 7 |
Hb_108886_010 |
0.2998736695 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_003025_070 |
0.3026824398 |
- |
- |
PREDICTED: uncharacterized protein LOC103419743 [Malus domestica] |
| 9 |
Hb_003777_010 |
0.3067325512 |
- |
- |
- |
| 10 |
Hb_003055_170 |
0.3130463691 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 25 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001338_060 |
0.3144037243 |
- |
- |
- |
| 12 |
Hb_171537_010 |
0.3182725464 |
- |
- |
serine/threonine-protein kinase bri1, putative [Ricinus communis] |
| 13 |
Hb_001387_030 |
0.3185514326 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0013s13920g [Populus trichocarpa] |
| 14 |
Hb_005045_040 |
0.3224621805 |
- |
- |
hypothetical protein POPTR_0017s04090g, partial [Populus trichocarpa] |
| 15 |
Hb_005699_080 |
0.3233833768 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Jatropha curcas] |
| 16 |
Hb_001462_020 |
0.3275617901 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 17 |
Hb_071586_010 |
0.3277035073 |
- |
- |
PREDICTED: uncharacterized protein LOC105632410 [Jatropha curcas] |
| 18 |
Hb_074108_010 |
0.3280459816 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 19 |
Hb_161072_010 |
0.3305408035 |
- |
- |
- |
| 20 |
Hb_007537_010 |
0.3325208027 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |