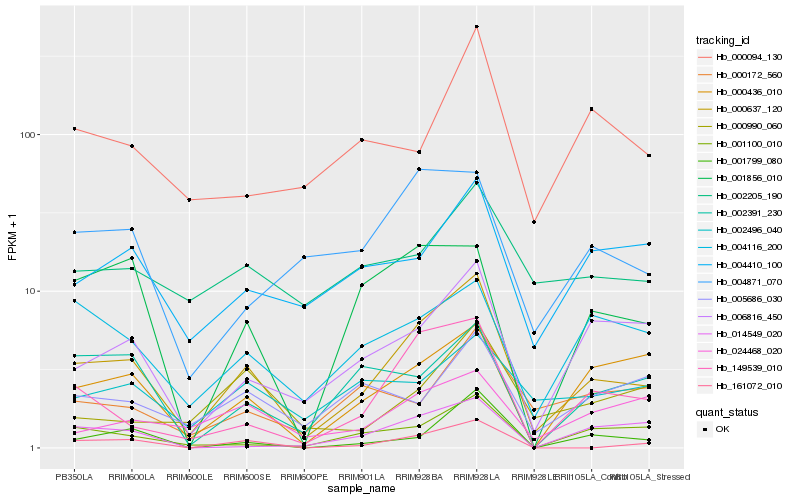
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000637_120 |
0.0 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 2 |
Hb_149539_010 |
0.2037564597 |
- |
- |
- |
| 3 |
Hb_006816_450 |
0.2046839758 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_161072_010 |
0.2067126004 |
- |
- |
- |
| 5 |
Hb_000436_010 |
0.2174291929 |
- |
- |
PREDICTED: probable polyamine transporter At1g31830 isoform X1 [Eucalyptus grandis] |
| 6 |
Hb_001100_010 |
0.2205404748 |
- |
- |
F-box/FBD/LRR-repeat protein [Medicago truncatula] |
| 7 |
Hb_001799_080 |
0.2287814389 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 8 |
Hb_004410_100 |
0.230709064 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 21 [Jatropha curcas] |
| 9 |
Hb_004116_200 |
0.234820262 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 10 |
Hb_004871_070 |
0.2382483326 |
- |
- |
PREDICTED: uncharacterized protein LOC105632531 [Jatropha curcas] |
| 11 |
Hb_024468_020 |
0.2393719366 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 12 |
Hb_000094_130 |
0.2424518164 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 13 |
Hb_001856_010 |
0.2433302388 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_002391_230 |
0.2438548315 |
- |
- |
hypothetical protein RCOM_0745080 [Ricinus communis] |
| 15 |
Hb_014549_020 |
0.2446936193 |
- |
- |
hypothetical protein POPTR_0013s12350g [Populus trichocarpa] |
| 16 |
Hb_002205_190 |
0.2492285959 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 17 |
Hb_000990_060 |
0.2495997639 |
- |
- |
PREDICTED: protein EMBRYONIC FLOWER 1 [Jatropha curcas] |
| 18 |
Hb_002496_040 |
0.2498437789 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03380, mitochondrial [Jatropha curcas] |
| 19 |
Hb_005686_030 |
0.2502034942 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03880, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_000172_560 |
0.2520694296 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750 [Jatropha curcas] |