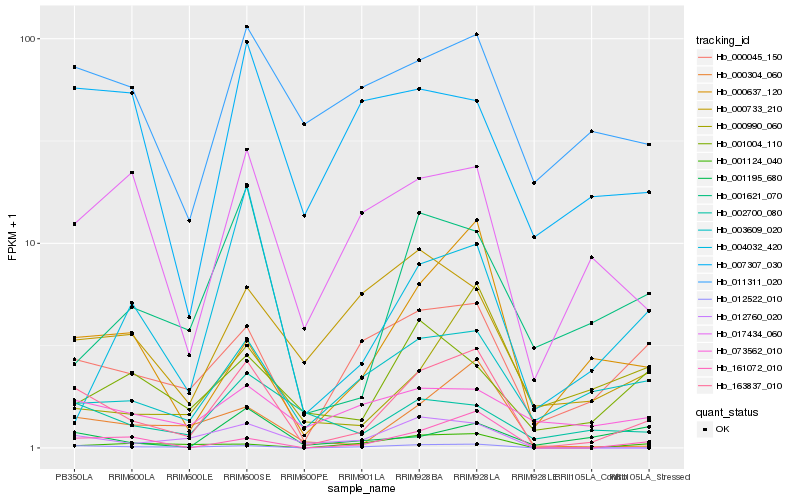
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_163837_010 |
0.0 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Eucalyptus grandis] |
| 2 |
Hb_012522_010 |
0.2397824309 |
- |
- |
hypothetical protein VITISV_003261 [Vitis vinifera] |
| 3 |
Hb_161072_010 |
0.2405004839 |
- |
- |
- |
| 4 |
Hb_000304_060 |
0.2429881512 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0010s15360g [Populus trichocarpa] |
| 5 |
Hb_000045_150 |
0.2528392415 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 6 |
Hb_000637_120 |
0.256951928 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 7 |
Hb_012760_020 |
0.2681595165 |
- |
- |
PREDICTED: uncharacterized protein LOC105647080 [Jatropha curcas] |
| 8 |
Hb_003609_020 |
0.2726294643 |
desease resistance |
Gene Name: Clusterin |
hypothetical protein CISIN_1g001897mg [Citrus sinensis] |
| 9 |
Hb_017434_060 |
0.2831393209 |
- |
- |
PREDICTED: 60S ribosomal export protein NMD3 [Jatropha curcas] |
| 10 |
Hb_001621_070 |
0.2840578795 |
- |
- |
hypothetical protein POPTR_0008s05890g [Populus trichocarpa] |
| 11 |
Hb_011311_020 |
0.2962699293 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002700_080 |
0.2987281336 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_24521 [Jatropha curcas] |
| 13 |
Hb_001124_040 |
0.2991866223 |
- |
- |
PREDICTED: uncharacterized protein LOC103413595 [Malus domestica] |
| 14 |
Hb_001195_680 |
0.2998927217 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 [Jatropha curcas] |
| 15 |
Hb_004032_420 |
0.3026492744 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 25-like [Jatropha curcas] |
| 16 |
Hb_000733_210 |
0.3043423897 |
- |
- |
putative serine/threonine protein phosphatase [Angiostrongylus costaricensis] |
| 17 |
Hb_007307_030 |
0.3066538942 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 18 |
Hb_073562_010 |
0.3080286599 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 19 |
Hb_000990_060 |
0.3087693769 |
- |
- |
PREDICTED: protein EMBRYONIC FLOWER 1 [Jatropha curcas] |
| 20 |
Hb_001004_110 |
0.3090040681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |