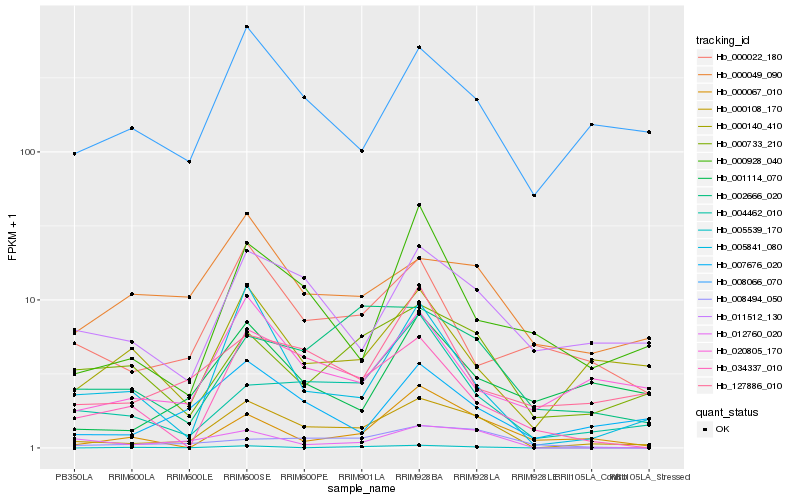
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000108_170 |
0.0 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 2 |
Hb_007676_020 |
0.2360758509 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_011512_130 |
0.2381224102 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At2g33170 [Jatropha curcas] |
| 4 |
Hb_005841_080 |
0.2400184644 |
- |
- |
hypothetical protein POPTR_0018s11370g [Populus trichocarpa] |
| 5 |
Hb_008066_070 |
0.2441522224 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 6 |
Hb_012760_020 |
0.2451314823 |
- |
- |
PREDICTED: uncharacterized protein LOC105647080 [Jatropha curcas] |
| 7 |
Hb_000733_210 |
0.246165723 |
- |
- |
putative serine/threonine protein phosphatase [Angiostrongylus costaricensis] |
| 8 |
Hb_002666_020 |
0.2511665831 |
- |
- |
VAMP/SYNAPTOBREVIN-ASSOCIATED protein 27-1 [Populus trichocarpa] |
| 9 |
Hb_004462_010 |
0.2535394384 |
- |
- |
PREDICTED: primary amine oxidase-like [Jatropha curcas] |
| 10 |
Hb_034337_010 |
0.2541970152 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 11 |
Hb_020805_170 |
0.2549797941 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 12 |
Hb_008494_050 |
0.2554334849 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 13 |
Hb_005539_170 |
0.2557183596 |
- |
- |
PREDICTED: uncharacterized protein LOC101502180 [Cicer arietinum] |
| 14 |
Hb_000049_090 |
0.2585003111 |
- |
- |
PREDICTED: uncharacterized protein LOC105644472 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000140_410 |
0.2636148266 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 16 |
Hb_127886_010 |
0.265407293 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 17 |
Hb_000022_180 |
0.2654775037 |
- |
- |
PREDICTED: RING finger and CHY zinc finger domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000928_040 |
0.2661979738 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_001114_070 |
0.2668944475 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At1g58400 [Jatropha curcas] |
| 20 |
Hb_000067_010 |
0.2682922757 |
- |
- |
hypothetical protein B456_002G147500 [Gossypium raimondii] |