| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
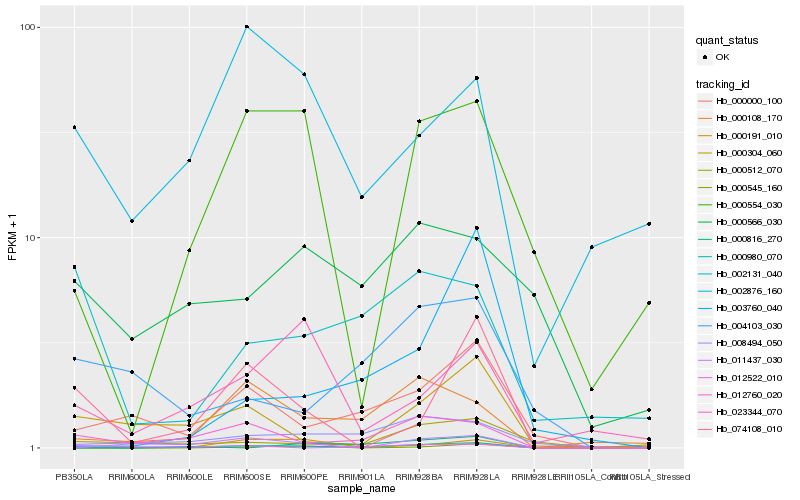
Hb_000980_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103440883 [Malus domestica] |
| 2 |
Hb_012760_020 |
0.2498577231 |
- |
- |
PREDICTED: uncharacterized protein LOC105647080 [Jatropha curcas] |
| 3 |
Hb_008494_050 |
0.2671873107 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 4 |
Hb_000000_100 |
0.2701560893 |
- |
- |
hypothetical protein POPTR_0002s13260g [Populus trichocarpa] |
| 5 |
Hb_000108_170 |
0.2711961643 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 6 |
Hb_000554_030 |
0.2819153966 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000304_060 |
0.3153444836 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0010s15360g [Populus trichocarpa] |
| 8 |
Hb_000191_010 |
0.3249741247 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 9 |
Hb_074108_010 |
0.3250662455 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 10 |
Hb_011437_030 |
0.3282813162 |
- |
- |
PREDICTED: uncharacterized protein LOC105037131, partial [Elaeis guineensis] |
| 11 |
Hb_000512_070 |
0.3290711113 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71250-like [Jatropha curcas] |
| 12 |
Hb_000545_160 |
0.333739858 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 13 |
Hb_023344_070 |
0.3356962242 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000816_270 |
0.3449816233 |
- |
- |
- |
| 15 |
Hb_003760_040 |
0.3452249618 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 16 |
Hb_002876_160 |
0.3480252287 |
- |
- |
sucrose transporter 1 [Hevea brasiliensis] |
| 17 |
Hb_000566_030 |
0.3485353478 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 18 |
Hb_002131_040 |
0.3494940959 |
- |
- |
proton-dependent oligopeptide transport family protein [Populus trichocarpa] |
| 19 |
Hb_004103_030 |
0.3497423626 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_012522_010 |
0.3541641569 |
- |
- |
hypothetical protein VITISV_003261 [Vitis vinifera] |