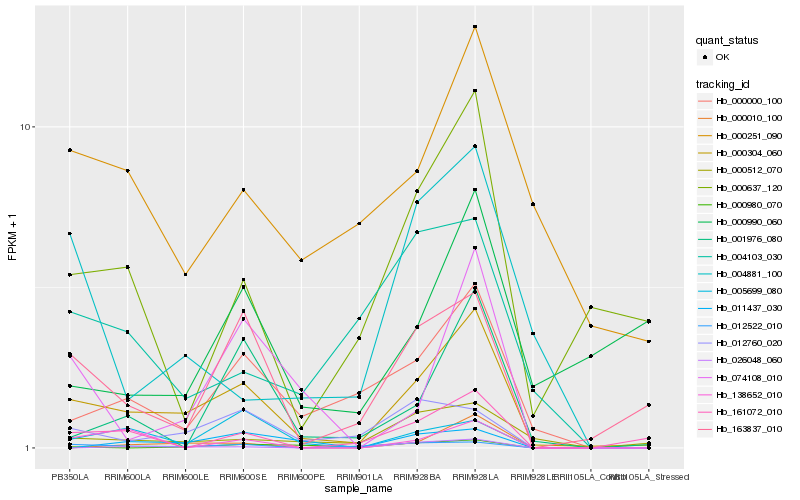
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000304_060 |
0.0 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0010s15360g [Populus trichocarpa] |
| 2 |
Hb_163837_010 |
0.2429881512 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Eucalyptus grandis] |
| 3 |
Hb_000000_100 |
0.24373526 |
- |
- |
hypothetical protein POPTR_0002s13260g [Populus trichocarpa] |
| 4 |
Hb_012522_010 |
0.2454728707 |
- |
- |
hypothetical protein VITISV_003261 [Vitis vinifera] |
| 5 |
Hb_012760_020 |
0.2691224179 |
- |
- |
PREDICTED: uncharacterized protein LOC105647080 [Jatropha curcas] |
| 6 |
Hb_000512_070 |
0.2693788093 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71250-like [Jatropha curcas] |
| 7 |
Hb_000251_090 |
0.2802933142 |
- |
- |
PREDICTED: two-pore potassium channel 3-like [Jatropha curcas] |
| 8 |
Hb_000637_120 |
0.280452409 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 9 |
Hb_161072_010 |
0.2812523778 |
- |
- |
- |
| 10 |
Hb_005699_080 |
0.2877090207 |
- |
- |
PREDICTED: ABC transporter G family member 10-like [Jatropha curcas] |
| 11 |
Hb_074108_010 |
0.288646596 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 12 |
Hb_001976_080 |
0.2953828997 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor AIL6 [Jatropha curcas] |
| 13 |
Hb_004103_030 |
0.3032606534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004881_100 |
0.306171528 |
- |
- |
Phosphoribosyltransferase family protein isoform 4 [Theobroma cacao] |
| 15 |
Hb_000980_070 |
0.3153444836 |
- |
- |
PREDICTED: uncharacterized protein LOC103440883 [Malus domestica] |
| 16 |
Hb_138652_010 |
0.3191900931 |
- |
- |
hypothetical protein JCGZ_13474 [Jatropha curcas] |
| 17 |
Hb_000010_100 |
0.3198913951 |
- |
- |
hypothetical protein POPTR_0016s14440g [Populus trichocarpa] |
| 18 |
Hb_000990_060 |
0.3242706702 |
- |
- |
PREDICTED: protein EMBRYONIC FLOWER 1 [Jatropha curcas] |
| 19 |
Hb_026048_060 |
0.3246976377 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 20 |
Hb_011437_030 |
0.3257261205 |
- |
- |
PREDICTED: uncharacterized protein LOC105037131, partial [Elaeis guineensis] |