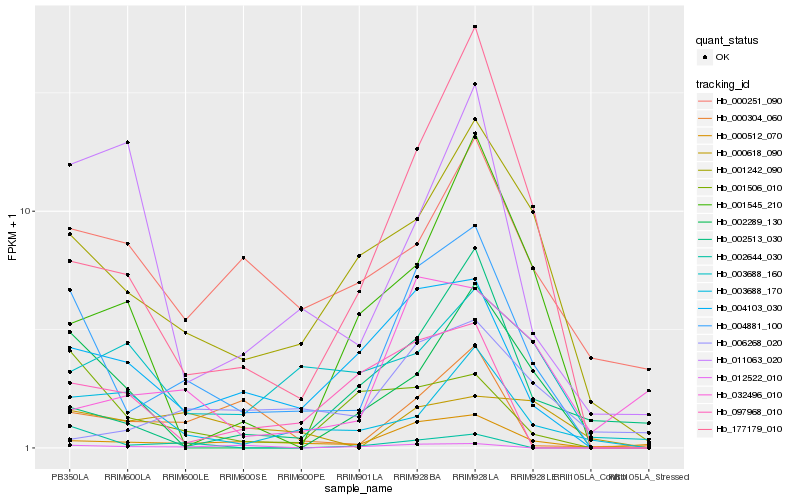
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004881_100 |
0.0 |
- |
- |
Phosphoribosyltransferase family protein isoform 4 [Theobroma cacao] |
| 2 |
Hb_000512_070 |
0.2277535809 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71250-like [Jatropha curcas] |
| 3 |
Hb_001242_090 |
0.2381322937 |
- |
- |
40S ribosomal protein S18 [Colletotrichum fioriniae PJ7] |
| 4 |
Hb_004103_030 |
0.2427183357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_177179_010 |
0.2588662142 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Populus euphratica] |
| 6 |
Hb_002289_130 |
0.2948693534 |
transcription factor |
TF Family: C2H2 |
arsenite inducuble RNA associated protein aip-1, putative [Ricinus communis] |
| 7 |
Hb_001506_010 |
0.3031662849 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 8 |
Hb_000304_060 |
0.306171528 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0010s15360g [Populus trichocarpa] |
| 9 |
Hb_003688_170 |
0.3096850469 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 isoform X2 [Cucumis sativus] |
| 10 |
Hb_000251_090 |
0.3130241387 |
- |
- |
PREDICTED: two-pore potassium channel 3-like [Jatropha curcas] |
| 11 |
Hb_032496_010 |
0.3205250602 |
- |
- |
hypothetical protein POPTR_0012s10850g [Populus trichocarpa] |
| 12 |
Hb_002513_030 |
0.3268758197 |
- |
- |
PREDICTED: alpha-L-fucosidase 2-like isoform X2 [Elaeis guineensis] |
| 13 |
Hb_097968_010 |
0.3276521256 |
- |
- |
hypothetical protein PRUPE_ppa001143mg [Prunus persica] |
| 14 |
Hb_001545_210 |
0.3307628084 |
- |
- |
hypothetical protein JCGZ_11132 [Jatropha curcas] |
| 15 |
Hb_006268_020 |
0.3366332006 |
- |
- |
hypothetical protein JCGZ_10516 [Jatropha curcas] |
| 16 |
Hb_011063_020 |
0.3368747274 |
- |
- |
PREDICTED: ribosomal L1 domain-containing protein 1-like [Jatropha curcas] |
| 17 |
Hb_002644_030 |
0.3415541185 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like, partial [Eucalyptus grandis] |
| 18 |
Hb_012522_010 |
0.3442627266 |
- |
- |
hypothetical protein VITISV_003261 [Vitis vinifera] |
| 19 |
Hb_003688_160 |
0.3450711666 |
- |
- |
Presenilin-like family protein [Populus trichocarpa] |
| 20 |
Hb_000618_090 |
0.3464434102 |
- |
- |
- |