| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
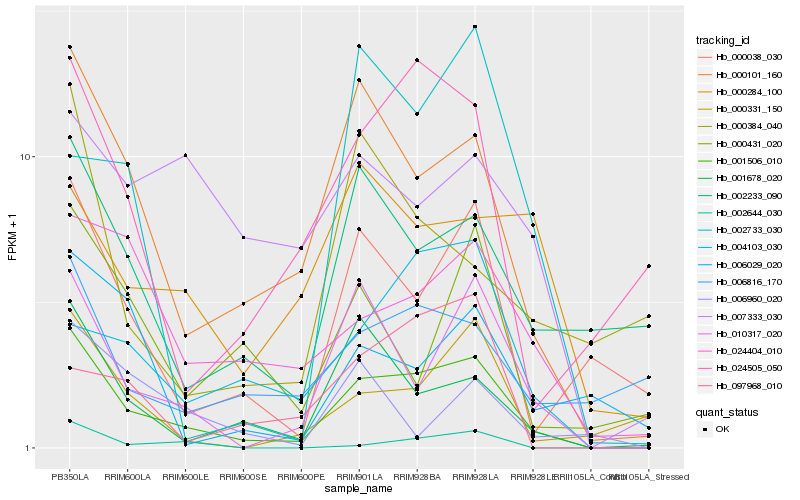
Hb_001506_010 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 2 |
Hb_000101_160 |
0.1820740125 |
- |
- |
PREDICTED: uncharacterized protein LOC105133607 isoform X1 [Populus euphratica] |
| 3 |
Hb_010317_020 |
0.2052234849 |
- |
- |
PREDICTED: sarcoplasmic reticulum histidine-rich calcium-binding protein [Jatropha curcas] |
| 4 |
Hb_001678_020 |
0.2241648774 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 5 |
Hb_000038_030 |
0.237824535 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7-like [Glycine max] |
| 6 |
Hb_024404_010 |
0.2443360273 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Vitis vinifera] |
| 7 |
Hb_024505_050 |
0.2516556299 |
transcription factor |
TF Family: ERF |
PPLZ02 family protein [Populus trichocarpa] |
| 8 |
Hb_004103_030 |
0.2534161709 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002233_090 |
0.255369446 |
- |
- |
PREDICTED: bromodomain-containing protein 3 [Jatropha curcas] |
| 10 |
Hb_000331_150 |
0.2565796735 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 11 |
Hb_006816_170 |
0.2667493304 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |
| 12 |
Hb_002644_030 |
0.267027416 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like, partial [Eucalyptus grandis] |
| 13 |
Hb_000431_020 |
0.2700758399 |
- |
- |
hypothetical protein JCGZ_06801 [Jatropha curcas] |
| 14 |
Hb_097968_010 |
0.282425001 |
- |
- |
hypothetical protein PRUPE_ppa001143mg [Prunus persica] |
| 15 |
Hb_000284_100 |
0.2840184058 |
- |
- |
PREDICTED: transcription factor GTE11 isoform X2 [Populus euphratica] |
| 16 |
Hb_006029_020 |
0.2842662781 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1, partial [Vitis vinifera] |
| 17 |
Hb_007333_030 |
0.2893398611 |
- |
- |
- |
| 18 |
Hb_000384_040 |
0.2923780585 |
- |
- |
PREDICTED: uncharacterized protein LOC105640093 [Jatropha curcas] |
| 19 |
Hb_002733_030 |
0.295824232 |
- |
- |
PREDICTED: uncharacterized protein LOC104248159 [Nicotiana sylvestris] |
| 20 |
Hb_006960_020 |
0.2963197382 |
- |
- |
PREDICTED: uncharacterized protein LOC102617255 [Citrus sinensis] |