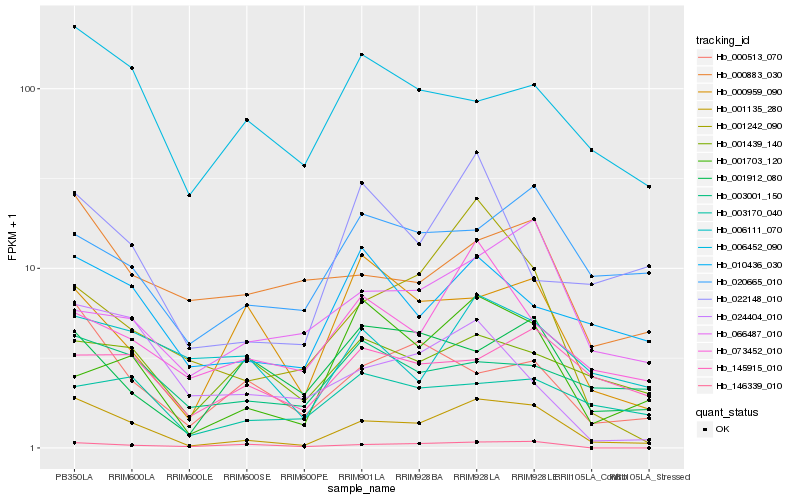
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001135_280 |
0.0 |
- |
- |
- |
| 2 |
Hb_000513_070 |
0.233277709 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 42 [Jatropha curcas] |
| 3 |
Hb_010436_030 |
0.2349812388 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 4 |
Hb_001242_090 |
0.2359055581 |
- |
- |
40S ribosomal protein S18 [Colletotrichum fioriniae PJ7] |
| 5 |
Hb_000883_030 |
0.2376475355 |
- |
- |
PREDICTED: uncharacterized protein LOC105636683 [Jatropha curcas] |
| 6 |
Hb_006111_070 |
0.2413650805 |
- |
- |
hypothetical protein POPTR_0008s07660g [Populus trichocarpa] |
| 7 |
Hb_006452_090 |
0.2423250078 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 5B [Jatropha curcas] |
| 8 |
Hb_073452_010 |
0.2423698521 |
- |
- |
PREDICTED: chaperonin CPN60-like 2, mitochondrial isoform X2 [Jatropha curcas] |
| 9 |
Hb_001703_120 |
0.2448778502 |
- |
- |
PREDICTED: uncharacterized protein LOC105116966 isoform X2 [Populus euphratica] |
| 10 |
Hb_020665_010 |
0.2503112177 |
- |
- |
PREDICTED: uncharacterized protein LOC103438977 isoform X2 [Malus domestica] |
| 11 |
Hb_001912_080 |
0.250640326 |
- |
- |
vacuolar protein sorting-associated protein, putative [Ricinus communis] |
| 12 |
Hb_022148_010 |
0.250661892 |
- |
- |
PREDICTED: uncharacterized protein LOC105638910 [Jatropha curcas] |
| 13 |
Hb_000959_090 |
0.2506974876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_146339_010 |
0.2511345418 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 15 |
Hb_003170_040 |
0.2522731609 |
- |
- |
PREDICTED: microfibrillar-associated protein 1-like [Prunus mume] |
| 16 |
Hb_145915_010 |
0.2525620322 |
- |
- |
PREDICTED: uncharacterized protein LOC105630963 [Jatropha curcas] |
| 17 |
Hb_066487_010 |
0.2526580509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_024404_010 |
0.2560096571 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Vitis vinifera] |
| 19 |
Hb_001439_140 |
0.2567471667 |
- |
- |
pattern formation protein, putative [Ricinus communis] |
| 20 |
Hb_003001_150 |
0.2593518188 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |