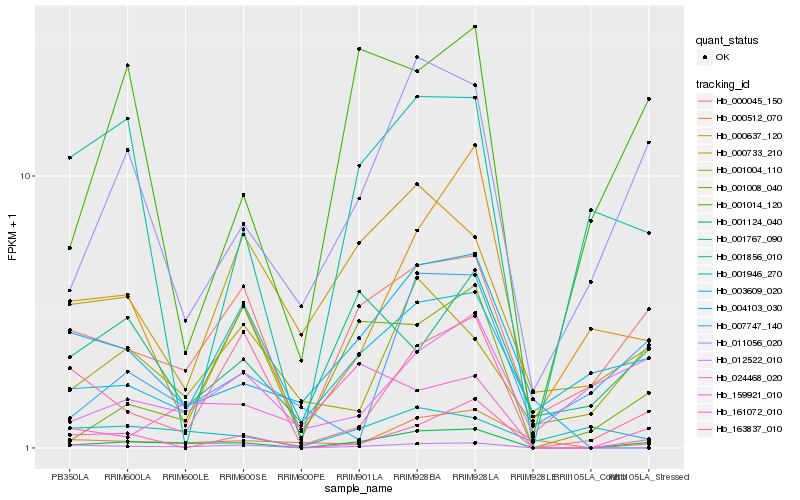
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001124_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103413595 [Malus domestica] |
| 2 |
Hb_011056_020 |
0.2311411129 |
transcription factor |
TF Family: ERF |
AP2 domain transcription factor RAP2.3, putative [Ricinus communis] |
| 3 |
Hb_000045_150 |
0.2374515219 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 4 |
Hb_161072_010 |
0.267982469 |
- |
- |
- |
| 5 |
Hb_012522_010 |
0.2795884481 |
- |
- |
hypothetical protein VITISV_003261 [Vitis vinifera] |
| 6 |
Hb_001014_120 |
0.2848787896 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor ERF113 [Jatropha curcas] |
| 7 |
Hb_001008_040 |
0.2856071115 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 8 |
Hb_000512_070 |
0.2882971414 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71250-like [Jatropha curcas] |
| 9 |
Hb_000637_120 |
0.2886952659 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 10 |
Hb_163837_010 |
0.2991866223 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Eucalyptus grandis] |
| 11 |
Hb_024468_020 |
0.3048150624 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 12 |
Hb_007747_140 |
0.304985347 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_003609_020 |
0.3052515923 |
desease resistance |
Gene Name: Clusterin |
hypothetical protein CISIN_1g001897mg [Citrus sinensis] |
| 14 |
Hb_000733_210 |
0.3055165333 |
- |
- |
putative serine/threonine protein phosphatase [Angiostrongylus costaricensis] |
| 15 |
Hb_004103_030 |
0.3070622067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001004_110 |
0.3081726251 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001767_090 |
0.3082257399 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 18 |
Hb_159921_010 |
0.308552309 |
- |
- |
- |
| 19 |
Hb_001946_270 |
0.3200565743 |
- |
- |
hypothetical protein JCGZ_13311 [Jatropha curcas] |
| 20 |
Hb_001856_010 |
0.3215466005 |
- |
- |
DNA binding protein, putative [Ricinus communis] |