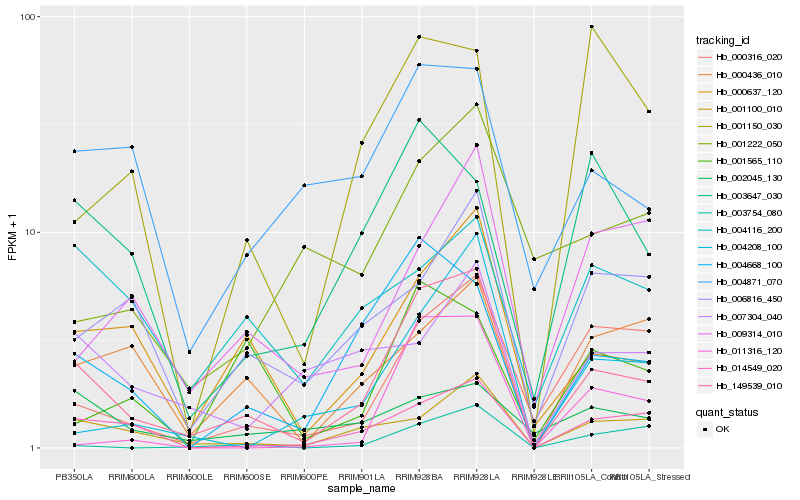
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_149539_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000637_120 |
0.2037564597 |
- |
- |
PREDICTED: aspartic proteinase Asp1 [Jatropha curcas] |
| 3 |
Hb_004668_100 |
0.2081706487 |
- |
- |
- |
| 4 |
Hb_014549_020 |
0.2114322219 |
- |
- |
hypothetical protein POPTR_0013s12350g [Populus trichocarpa] |
| 5 |
Hb_001100_010 |
0.2117248295 |
- |
- |
F-box/FBD/LRR-repeat protein [Medicago truncatula] |
| 6 |
Hb_000316_020 |
0.2416192602 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 7 |
Hb_003754_080 |
0.2556554025 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 8 |
Hb_004208_100 |
0.2591913679 |
- |
- |
Ribosomal protein L3 plastid isoform 1 [Theobroma cacao] |
| 9 |
Hb_004871_070 |
0.2598926276 |
- |
- |
PREDICTED: uncharacterized protein LOC105632531 [Jatropha curcas] |
| 10 |
Hb_000436_010 |
0.2640553106 |
- |
- |
PREDICTED: probable polyamine transporter At1g31830 isoform X1 [Eucalyptus grandis] |
| 11 |
Hb_006816_450 |
0.2676149617 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_011316_120 |
0.2700994606 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH118-like [Jatropha curcas] |
| 13 |
Hb_003647_030 |
0.2746029258 |
- |
- |
PREDICTED: probable glutathione S-transferase [Populus euphratica] |
| 14 |
Hb_007304_040 |
0.279041233 |
- |
- |
PREDICTED: polyadenylate-binding protein 3 [Jatropha curcas] |
| 15 |
Hb_001565_110 |
0.283226125 |
- |
- |
PREDICTED: putative ABC transporter B family member 8 [Jatropha curcas] |
| 16 |
Hb_009314_010 |
0.283506541 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 17 |
Hb_004116_200 |
0.2849918332 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase [Jatropha curcas] |
| 18 |
Hb_002045_130 |
0.2858045849 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 19 |
Hb_001150_030 |
0.2877650701 |
- |
- |
NADP-dependent oxidoreductase P1 isoform 3 [Theobroma cacao] |
| 20 |
Hb_001222_050 |
0.288573922 |
- |
- |
hypothetical protein CICLE_v100161911mg, partial [Citrus clementina] |