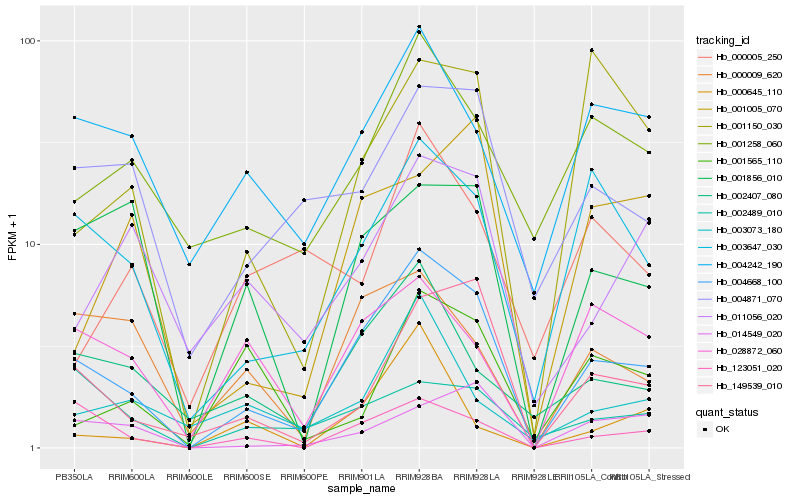
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004668_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_149539_010 |
0.2081706487 |
- |
- |
- |
| 3 |
Hb_003647_030 |
0.2123812186 |
- |
- |
PREDICTED: probable glutathione S-transferase [Populus euphratica] |
| 4 |
Hb_123051_020 |
0.2366811941 |
- |
- |
hypothetical protein POPTR_2151s00200g [Populus trichocarpa] |
| 5 |
Hb_002407_080 |
0.2377082531 |
- |
- |
hypothetical protein JCGZ_00752 [Jatropha curcas] |
| 6 |
Hb_004871_070 |
0.2399834442 |
- |
- |
PREDICTED: uncharacterized protein LOC105632531 [Jatropha curcas] |
| 7 |
Hb_001258_060 |
0.2424582225 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxykinase [ATP] [Jatropha curcas] |
| 8 |
Hb_004242_190 |
0.2467194922 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 9 |
Hb_001856_010 |
0.2528629312 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_014549_020 |
0.2574044834 |
- |
- |
hypothetical protein POPTR_0013s12350g [Populus trichocarpa] |
| 11 |
Hb_000009_620 |
0.259843717 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_001150_030 |
0.2624728936 |
- |
- |
NADP-dependent oxidoreductase P1 isoform 3 [Theobroma cacao] |
| 13 |
Hb_011056_020 |
0.2628048185 |
transcription factor |
TF Family: ERF |
AP2 domain transcription factor RAP2.3, putative [Ricinus communis] |
| 14 |
Hb_028872_060 |
0.2640695785 |
- |
- |
PREDICTED: ankyrin-3-like [Jatropha curcas] |
| 15 |
Hb_003073_180 |
0.2683576756 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 16 |
Hb_000645_110 |
0.2833280271 |
transcription factor |
TF Family: GRAS |
PREDICTED: DELLA protein RGL1-like [Jatropha curcas] |
| 17 |
Hb_001565_110 |
0.2852213599 |
- |
- |
PREDICTED: putative ABC transporter B family member 8 [Jatropha curcas] |
| 18 |
Hb_001005_070 |
0.2858265533 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF113-like [Jatropha curcas] |
| 19 |
Hb_002489_010 |
0.2866655725 |
- |
- |
hypothetical protein JCGZ_17199 [Jatropha curcas] |
| 20 |
Hb_000005_250 |
0.2869276262 |
- |
- |
conserved hypothetical protein [Ricinus communis] |