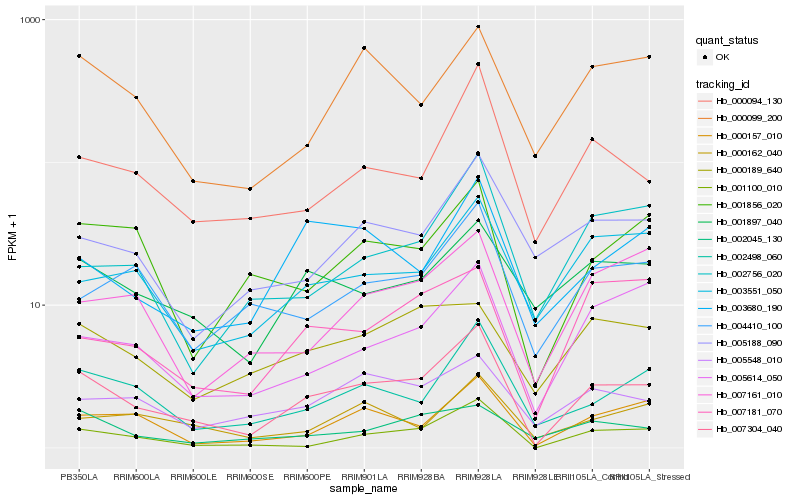
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007304_040 |
0.0 |
- |
- |
PREDICTED: polyadenylate-binding protein 3 [Jatropha curcas] |
| 2 |
Hb_001100_010 |
0.1634837647 |
- |
- |
F-box/FBD/LRR-repeat protein [Medicago truncatula] |
| 3 |
Hb_002498_060 |
0.1678027236 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_000094_130 |
0.1701375016 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 5 |
Hb_000157_010 |
0.1778353479 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 6 |
Hb_005614_050 |
0.188061798 |
- |
- |
PREDICTED: aspartate aminotransferase, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_003680_190 |
0.1883346574 |
- |
- |
hypothetical protein POPTR_0006s11880g [Populus trichocarpa] |
| 8 |
Hb_001897_040 |
0.1901991536 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 9 |
Hb_005548_010 |
0.1919909869 |
- |
- |
PREDICTED: DNA repair protein RAD51 homolog 2 [Jatropha curcas] |
| 10 |
Hb_001856_020 |
0.192633384 |
transcription factor |
TF Family: Orphans |
ethylene receptor, putative [Ricinus communis] |
| 11 |
Hb_003551_050 |
0.1941241324 |
- |
- |
PREDICTED: mitochondrial fission 1 protein A [Jatropha curcas] |
| 12 |
Hb_002045_130 |
0.1951013663 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 13 |
Hb_005188_090 |
0.1951694978 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase gamma 7-like [Jatropha curcas] |
| 14 |
Hb_000099_200 |
0.1952768078 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004410_100 |
0.197532632 |
transcription factor |
TF Family: Orphans |
PREDICTED: B-box zinc finger protein 21 [Jatropha curcas] |
| 16 |
Hb_007181_070 |
0.198101203 |
- |
- |
PREDICTED: alpha-galactosidase-like [Jatropha curcas] |
| 17 |
Hb_000162_040 |
0.199717003 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |
| 18 |
Hb_002756_020 |
0.2032198906 |
transcription factor |
TF Family: HB |
bel1 homeotic protein, putative [Ricinus communis] |
| 19 |
Hb_007161_010 |
0.2034581043 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000189_640 |
0.204367971 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |