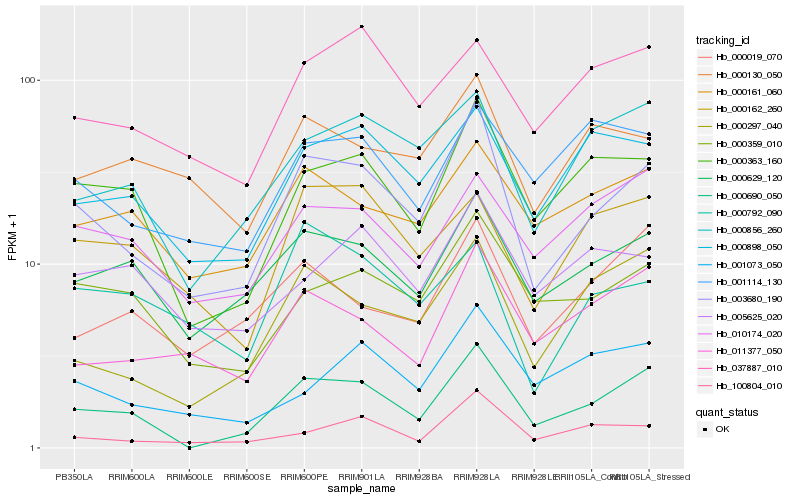
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003680_190 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s11880g [Populus trichocarpa] |
| 2 |
Hb_000690_050 |
0.1305620375 |
- |
- |
- |
| 3 |
Hb_000629_120 |
0.1436106641 |
- |
- |
Polynucleotidyl transferase, ribonuclease H-like superfamily protein [Theobroma cacao] |
| 4 |
Hb_000363_160 |
0.1471058143 |
- |
- |
Heterogeneous nuclear ribonucleoprotein 27C, putative [Ricinus communis] |
| 5 |
Hb_000359_010 |
0.1535141336 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 6 |
Hb_000898_050 |
0.1619866788 |
- |
- |
- |
| 7 |
Hb_000297_040 |
0.1629634863 |
- |
- |
pre-mRNA cleavage factor im, 25kD subunit, putative [Ricinus communis] |
| 8 |
Hb_100804_010 |
0.1630489478 |
- |
- |
BAHD family acyltransferase, clade V, putative [Theobroma cacao] |
| 9 |
Hb_000856_260 |
0.1630902981 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0015s07640g [Populus trichocarpa] |
| 10 |
Hb_011377_050 |
0.1634219187 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At3g02290-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000161_060 |
0.1665936904 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 [Jatropha curcas] |
| 12 |
Hb_000130_050 |
0.1671197051 |
- |
- |
14-3-3 protein, putative [Ricinus communis] |
| 13 |
Hb_001073_050 |
0.174993722 |
- |
- |
- |
| 14 |
Hb_000162_260 |
0.1759876585 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-1 [Nelumbo nucifera] |
| 15 |
Hb_000019_070 |
0.1762726289 |
- |
- |
PREDICTED: uncharacterized protein LOC105630200 [Jatropha curcas] |
| 16 |
Hb_005625_020 |
0.1776257207 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_037887_010 |
0.177851561 |
- |
- |
Charged multivesicular body protein 2a, putative [Ricinus communis] |
| 18 |
Hb_000792_090 |
0.1790695758 |
- |
- |
3-beta-hydroxy-delta5-steroid dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_010174_020 |
0.1794962945 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_001114_130 |
0.1796505604 |
transcription factor |
TF Family: MIKC |
PREDICTED: MADS-box protein SVP-like isoform X1 [Jatropha curcas] |