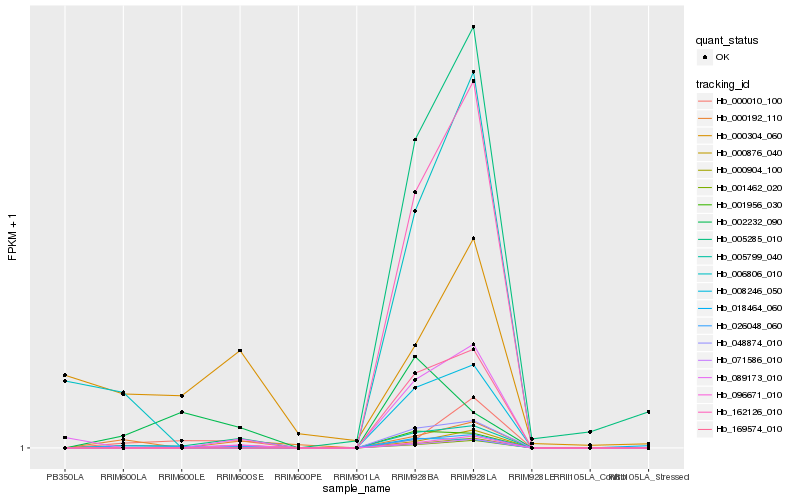
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026048_060 |
0.0 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 2 |
Hb_000010_100 |
0.2337962531 |
- |
- |
hypothetical protein POPTR_0016s14440g [Populus trichocarpa] |
| 3 |
Hb_000192_110 |
0.2822381601 |
- |
- |
PREDICTED: uncharacterized protein LOC105646856 [Jatropha curcas] |
| 4 |
Hb_000876_040 |
0.2873613073 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit B-like [Jatropha curcas] |
| 5 |
Hb_001956_030 |
0.287874741 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Populus euphratica] |
| 6 |
Hb_096671_010 |
0.290088291 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 7 |
Hb_000904_100 |
0.2902870517 |
- |
- |
hypothetical protein JCGZ_03883 [Jatropha curcas] |
| 8 |
Hb_071586_010 |
0.3008290301 |
- |
- |
PREDICTED: uncharacterized protein LOC105632410 [Jatropha curcas] |
| 9 |
Hb_001462_020 |
0.30087301 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 10 |
Hb_089173_010 |
0.3037582644 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 11 |
Hb_006806_010 |
0.3080127584 |
- |
- |
- |
| 12 |
Hb_018464_060 |
0.3172030875 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_005285_010 |
0.3196521707 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002232_090 |
0.319652406 |
- |
- |
STS14 protein precursor, putative [Ricinus communis] |
| 15 |
Hb_008246_050 |
0.320907453 |
- |
- |
- |
| 16 |
Hb_005799_040 |
0.32090966 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4-like isoform X1 [Gossypium raimondii] |
| 17 |
Hb_048874_010 |
0.3212647592 |
- |
- |
hypothetical protein POPTR_0016s10870g [Populus trichocarpa] |
| 18 |
Hb_169574_010 |
0.3214398656 |
- |
- |
PREDICTED: uncharacterized protein LOC104120373 [Nicotiana tomentosiformis] |
| 19 |
Hb_162126_010 |
0.3234476952 |
- |
- |
- |
| 20 |
Hb_000304_060 |
0.3246976377 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0010s15360g [Populus trichocarpa] |