| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
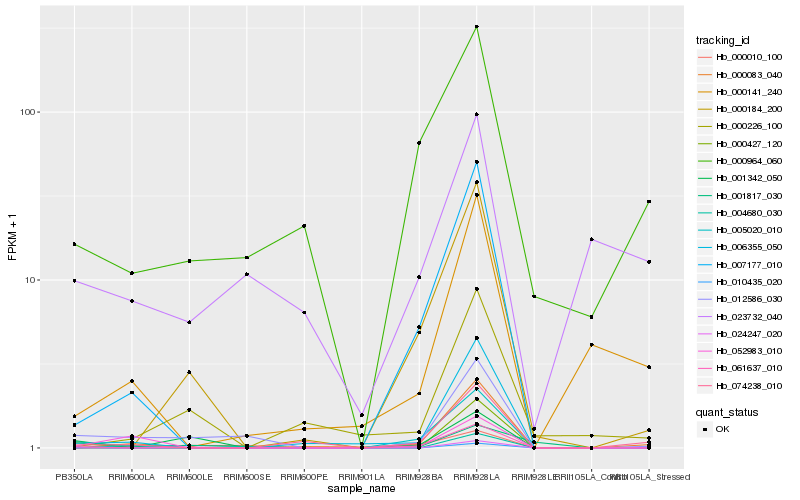
Hb_012586_030 |
0.0 |
- |
- |
hypothetical protein CICLE_v10014868mg [Citrus clementina] |
| 2 |
Hb_000010_100 |
0.246123212 |
- |
- |
hypothetical protein POPTR_0016s14440g [Populus trichocarpa] |
| 3 |
Hb_006355_050 |
0.2987979056 |
- |
- |
- |
| 4 |
Hb_001817_030 |
0.3029684868 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 5 |
Hb_001342_050 |
0.3030711345 |
- |
- |
- |
| 6 |
Hb_007177_010 |
0.3033452524 |
- |
- |
- |
| 7 |
Hb_061637_010 |
0.3036996394 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 8 |
Hb_000964_060 |
0.3089386151 |
- |
- |
hypothetical protein POPTR_0009s06660g [Populus trichocarpa] |
| 9 |
Hb_000427_120 |
0.3133940487 |
- |
- |
- |
| 10 |
Hb_000083_040 |
0.3155194473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_074238_010 |
0.3165797562 |
- |
- |
PREDICTED: uncharacterized protein LOC105797726 [Gossypium raimondii] |
| 12 |
Hb_004680_030 |
0.3179969512 |
- |
- |
PREDICTED: uncharacterized protein LOC105795864 [Gossypium raimondii] |
| 13 |
Hb_000184_200 |
0.320367854 |
- |
- |
- |
| 14 |
Hb_010435_020 |
0.3223132711 |
- |
- |
PREDICTED: uncharacterized protein LOC104216897 [Nicotiana sylvestris] |
| 15 |
Hb_023732_040 |
0.3250602681 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_052983_010 |
0.3251740104 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 17 |
Hb_005020_010 |
0.3271475213 |
- |
- |
- |
| 18 |
Hb_024247_020 |
0.3313372355 |
- |
- |
PREDICTED: uncharacterized protein LOC105634579 [Jatropha curcas] |
| 19 |
Hb_000226_100 |
0.334675387 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 20 |
Hb_000141_240 |
0.3359225745 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 6.3 isoform X1 [Jatropha curcas] |