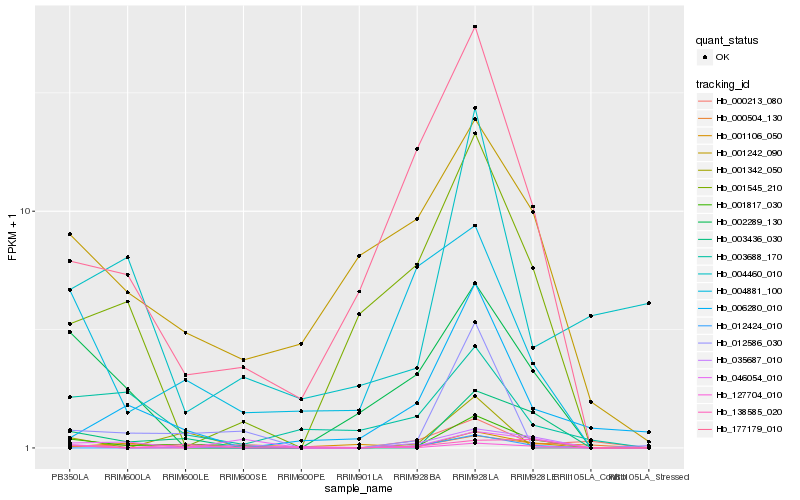
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001817_030 |
0.0 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 2 |
Hb_003436_030 |
0.2309037187 |
- |
- |
plant origin recognition complex subunit, putative [Ricinus communis] |
| 3 |
Hb_012586_030 |
0.3029684868 |
- |
- |
hypothetical protein CICLE_v10014868mg [Citrus clementina] |
| 4 |
Hb_177179_010 |
0.3145190807 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Populus euphratica] |
| 5 |
Hb_035687_010 |
0.3169173776 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 6 |
Hb_002289_130 |
0.3332278178 |
transcription factor |
TF Family: C2H2 |
arsenite inducuble RNA associated protein aip-1, putative [Ricinus communis] |
| 7 |
Hb_046054_010 |
0.3365663061 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 8 |
Hb_001242_090 |
0.3375795744 |
- |
- |
40S ribosomal protein S18 [Colletotrichum fioriniae PJ7] |
| 9 |
Hb_001545_210 |
0.3394351812 |
- |
- |
hypothetical protein JCGZ_11132 [Jatropha curcas] |
| 10 |
Hb_127704_010 |
0.3458625913 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_001106_050 |
0.349091574 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_004881_100 |
0.3492964289 |
- |
- |
Phosphoribosyltransferase family protein isoform 4 [Theobroma cacao] |
| 13 |
Hb_001342_050 |
0.3569791669 |
- |
- |
- |
| 14 |
Hb_138585_020 |
0.3589314302 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_006280_010 |
0.3608800591 |
transcription factor |
TF Family: ERF |
PREDICTED: AP2-like ethylene-responsive transcription factor TOE3 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003688_170 |
0.3632689329 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 isoform X2 [Cucumis sativus] |
| 17 |
Hb_012424_010 |
0.3653180978 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 18 |
Hb_000213_080 |
0.3656288165 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1-like [Vitis vinifera] |
| 19 |
Hb_004460_010 |
0.3668114649 |
- |
- |
PREDICTED: SKI/DACH domain-containing protein 1-like [Jatropha curcas] |
| 20 |
Hb_000504_130 |
0.3708411178 |
- |
- |
GPI transamidase component family protein [Populus trichocarpa] |